Metagenomics explores the collective genetic material of microbial communities directly from environmental samples, bypassing the need for culturing individual species. This powerful approach provides insights into the diversity, functions, and interactions of microorganisms in ecosystems ranging from soil to the human gut. Discover how metagenomics can revolutionize your understanding of microbial life by reading the full article.
Table of Comparison
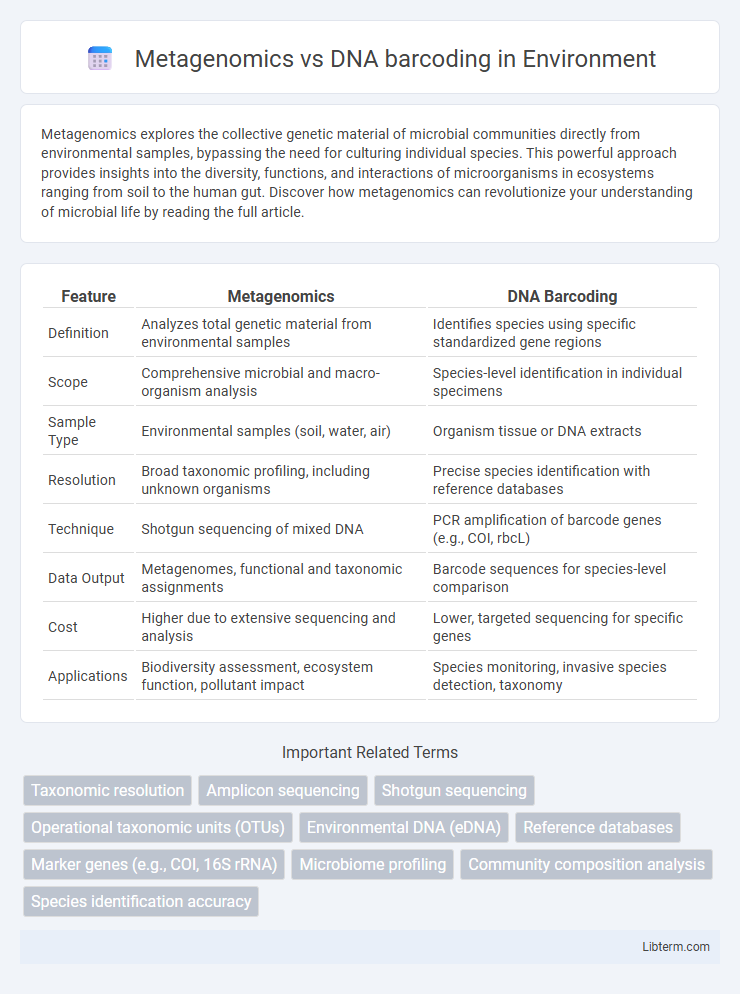
| Feature | Metagenomics | DNA Barcoding |
|---|---|---|
| Definition | Analyzes total genetic material from environmental samples | Identifies species using specific standardized gene regions |
| Scope | Comprehensive microbial and macro-organism analysis | Species-level identification in individual specimens |
| Sample Type | Environmental samples (soil, water, air) | Organism tissue or DNA extracts |
| Resolution | Broad taxonomic profiling, including unknown organisms | Precise species identification with reference databases |
| Technique | Shotgun sequencing of mixed DNA | PCR amplification of barcode genes (e.g., COI, rbcL) |
| Data Output | Metagenomes, functional and taxonomic assignments | Barcode sequences for species-level comparison |
| Cost | Higher due to extensive sequencing and analysis | Lower, targeted sequencing for specific genes |
| Applications | Biodiversity assessment, ecosystem function, pollutant impact | Species monitoring, invasive species detection, taxonomy |
Introduction to Metagenomics and DNA Barcoding
Metagenomics involves the comprehensive analysis of genetic material recovered directly from environmental samples, enabling the study of entire microbial communities without the need for cultivation. DNA barcoding uses short, standardized gene regions, such as the mitochondrial COI gene in animals, to identify and classify individual species with high precision. Both techniques revolutionize biodiversity assessment, with metagenomics offering broad community insights and DNA barcoding providing species-level resolution.
Fundamental Principles of Metagenomics
Metagenomics involves the analysis of genetic material recovered directly from environmental samples, enabling the study of entire microbial communities without the need for culturing individual species. It relies on high-throughput sequencing technologies to capture the collective genome (metagenome) and assess functional and taxonomic diversity. Unlike DNA barcoding, which targets specific genetic markers such as the mitochondrial COI gene for species identification, metagenomics provides a comprehensive view of microbial ecology by examining all genetic content simultaneously.
Core Concepts of DNA Barcoding
DNA barcoding relies on sequencing a short, standardized region of the genome, typically the mitochondrial COI gene in animals, to accurately identify species based on unique genetic markers. This method focuses on comparing the barcode sequence against a reference database to assign taxonomic identity with high precision. Unlike metagenomics, which analyzes entire genomic content from environmental samples, DNA barcoding provides targeted species identification and is widely used for biodiversity assessment and species authentication.
Methodological Differences
Metagenomics involves sequencing the entire genetic material from environmental samples, enabling comprehensive analysis of microbial communities without prior knowledge of organisms present. DNA barcoding targets specific gene regions, such as the mitochondrial COI gene in animals, to identify individual species by comparing sequences to reference databases. Metagenomics offers broader taxonomic resolution and functional insights, whereas DNA barcoding provides faster, cost-effective species identification with limited community context.
Applications in Biodiversity Assessment
Metagenomics enables comprehensive biodiversity assessment by analyzing environmental DNA from complex samples, revealing both known and unknown species across diverse ecosystems. DNA barcoding targets specific genetic markers to rapidly identify species, making it ideal for monitoring taxa with established reference libraries. Combining metagenomics and DNA barcoding enhances biodiversity evaluation by integrating broad community profiling with precise species identification.
Advantages and Limitations of Each Approach
Metagenomics enables comprehensive analysis of entire microbial communities by sequencing all genetic material, providing insights into functional potential and diversity beyond individual species; however, it requires advanced bioinformatics and can be cost-prohibitive. DNA barcoding offers rapid, cost-effective species identification through standardized genetic markers, making it suitable for biodiversity assessments but is limited in detecting rare or novel species and lacks functional information. Both methods face challenges with incomplete reference databases, impacting accuracy and taxonomic resolution in ecological studies.
Data Analysis and Interpretation
Metagenomics involves analyzing entire genetic material from environmental samples, enabling comprehensive data sets that capture microbial diversity but require complex bioinformatics tools for sequence assembly, annotation, and abundance estimation. DNA barcoding focuses on specific genetic markers, producing targeted data sets that simplify species identification but may miss broader ecological context and microbial interactions. Interpreting metagenomic data demands expertise in computational pipelines and statistical models to manage vast sequence variations, while DNA barcoding analysis relies on curated reference databases and sequence alignment algorithms for precise taxonomic classification.
Technological Requirements and Cost
Metagenomics requires high-throughput sequencing platforms and sophisticated bioinformatics tools to analyze complex microbial communities, leading to higher initial investment and operational costs compared to DNA barcoding. DNA barcoding typically employs targeted PCR amplification followed by Sanger sequencing, which demands less advanced infrastructure and is more cost-effective for species identification of individual organisms. Despite the lower cost of DNA barcoding, metagenomics provides comprehensive insights into microbial diversity and function, justifying its higher technological and financial requirements in ecological and environmental research.
Case Studies: Comparative Insights
Metagenomics enables comprehensive analysis of entire microbial communities in environmental samples, as demonstrated in a case study on ocean microbiomes revealing novel bacterial functions. DNA barcoding provides precise species-level identification, evidenced by rainforest biodiversity assessments accurately cataloging insect species diversity. Comparative insights show metagenomics excels in functional potential and community dynamics, while DNA barcoding offers species resolution, making their combination powerful for ecosystem and biodiversity studies.
Future Perspectives in Molecular Ecology
Metagenomics offers comprehensive insights into microbial community function and diversity by sequencing entire genomes from environmental samples, surpassing DNA barcoding which targets specific marker genes for species identification. Future perspectives in molecular ecology emphasize integrating metagenomics with advanced bioinformatics and machine learning to enhance ecosystem monitoring, biodiversity assessment, and understanding of microbial interactions. The continued development of high-throughput sequencing technologies and improved reference databases will drive more accurate, scalable, and rapid environmental DNA analyses critical for conservation and ecological forecasting.
Metagenomics Infographic
 libterm.com
libterm.com