Analog technology processes information through continuous signals, offering a natural and often more detailed representation of data compared to digital systems. This approach remains essential in audio recording, radio transmission, and vintage electronics, where signal fidelity and warmth are valued. Discover how understanding analog can enhance your appreciation of modern and classic technologies by reading the rest of the article.
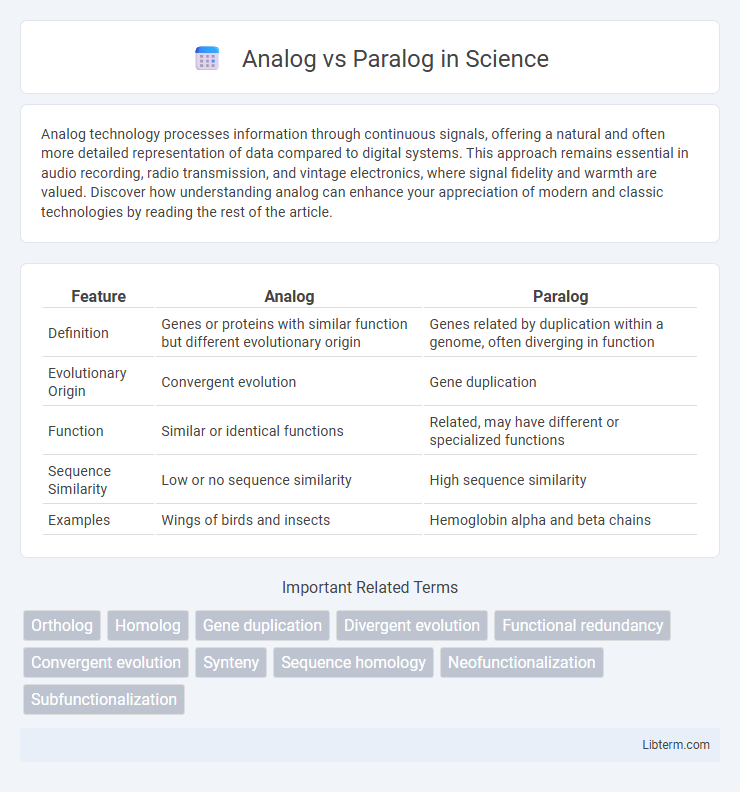
Table of Comparison
| Feature | Analog | Paralog |
|---|---|---|
| Definition | Genes or proteins with similar function but different evolutionary origin | Genes related by duplication within a genome, often diverging in function |
| Evolutionary Origin | Convergent evolution | Gene duplication |
| Function | Similar or identical functions | Related, may have different or specialized functions |
| Sequence Similarity | Low or no sequence similarity | High sequence similarity |
| Examples | Wings of birds and insects | Hemoglobin alpha and beta chains |
Understanding Analogs and Paralogs
Analogs are genes or proteins in different species that perform similar functions despite having different evolutionary origins, while paralogs arise from gene duplication events within the same species and often evolve new functions. Understanding analogs helps identify convergent evolution where distinct genetic paths result in similar biological roles, whereas studying paralogs provides insights into functional diversification and genomic innovation. Detailed analysis of sequence similarity, structural domains, and evolutionary lineage is essential to differentiate between analogs and paralogs accurately.
Evolutionary Origins of Analog vs Paralog Genes
Analog genes arise from convergent evolution where unrelated species independently develop similar traits or functions due to comparable environmental pressures, despite lacking a common ancestral gene. Paralog genes result from gene duplication events within the same genome, leading to gene families that diverge and acquire new or specialized functions over time. Understanding the evolutionary origins of analog and paralog genes illuminates mechanisms driving genetic diversity and adaptation across species.
Structural Differences Between Analogs and Paralogs
Analogs exhibit functional similarity but differ significantly in structural features due to convergent evolution, often lacking homologous protein domains. Paralogs arise from gene duplication events, maintaining conserved structural motifs and domain architectures despite evolving new functions. These structural differences between analogs and paralogs are critical for understanding evolutionary relationships and protein function diversification.
Functional Implications in Biological Systems
Analog proteins exhibit similar functions despite lacking common ancestry, often arising through convergent evolution, which enables different organisms to independently develop comparable traits. Paralog proteins result from gene duplication within the same organism, frequently diversifying to acquire novel or specialized functions that enhance biological complexity. Understanding the functional implications of analogs and paralogs informs evolutionary biology, protein engineering, and therapeutic target identification.
Methods for Identifying Analogs and Paralogs
Methods for identifying analogs primarily rely on structural and functional similarities despite low sequence homology, utilizing techniques such as protein structure comparison, conserved domain analysis, and functional assays. Paralogs are typically identified through sequence homology, phylogenetic analysis, and gene duplication events detected by bioinformatics tools like BLAST, OrthoFinder, and phylogenetic tree reconstruction. Combining multiple approaches, including sequence alignment, evolutionary context, and functional characterization, enhances accuracy in distinguishing analogs from paralogs.
Role in Gene Family Expansion
Analogs arise from convergent evolution, performing similar functions in different species without common ancestry, whereas paralogs originate from gene duplication events within a species and contribute directly to gene family expansion. Paralogs diversify through mutations, enabling new functions or subfunctions and increasing genetic complexity. This gene duplication and divergence process is fundamental for adaptive evolution and innovation within gene families.
Significance in Comparative Genomics
Analogous genes arise from convergent evolution and perform similar functions without shared ancestry, while paralogous genes result from gene duplication events within the same genome and often diverge to acquire new functions. Identifying paralogs is crucial in comparative genomics for understanding gene family expansions, functional diversification, and evolutionary innovation within species. Differentiating paralogs from analogs aids in reconstructing accurate phylogenies and elucidating mechanisms of molecular evolution.
Applications in Drug Discovery and Biotechnology
Analogs and paralogs play distinct roles in drug discovery and biotechnology; analogs are chemically modified compounds designed to mimic the structure and function of lead molecules, enabling the optimization of drug efficacy and safety profiles. Paralogs, which are gene copies resulting from duplication events with conserved but divergent functions, serve as critical targets for understanding protein families, facilitating the identification of selective inhibitors and reducing off-target effects in therapeutic development. Leveraging analogs accelerates lead optimization, while studying paralogs aids in deciphering functional diversity and specificity, integral for precision medicine and biotechnological innovation.
Challenges in Distinguishing Analogs from Paralogs
Distinguishing analogs from paralogs poses significant challenges due to functional convergence and sequence similarity resulting from evolutionary pressures rather than shared ancestry. Analytical methods must carefully assess structural, functional, and phylogenetic data to avoid misclassifying genes or proteins that perform similar roles but originate from distinct evolutionary events. Ambiguities in gene duplication and horizontal gene transfer further complicate the accurate identification of paralogs versus analogs in complex genomes.
Future Directions in Analog and Paralog Research
Future directions in analog and paralog research emphasize leveraging advanced genomic sequencing and bioinformatics tools to elucidate evolutionary relationships and functional divergence. Integrating multi-omics approaches permits detailed characterization of analogs and paralogs, aiding in drug target identification and synthetic biology applications. Enhanced computational modeling and machine learning algorithms are poised to accelerate discovery of novel paralogous gene functions, driving personalized medicine and adaptive biotechnology innovations.
Analog Infographic
 libterm.com
libterm.com