Ohnologs are gene duplicates originating from whole genome duplication events, playing a crucial role in evolutionary innovation and complexity within vertebrates. These duplicated genes often retain essential functions or evolve new roles, impacting development and disease. Explore this article to understand how ohnologs influence genome evolution and your biological blueprint.
Table of Comparison
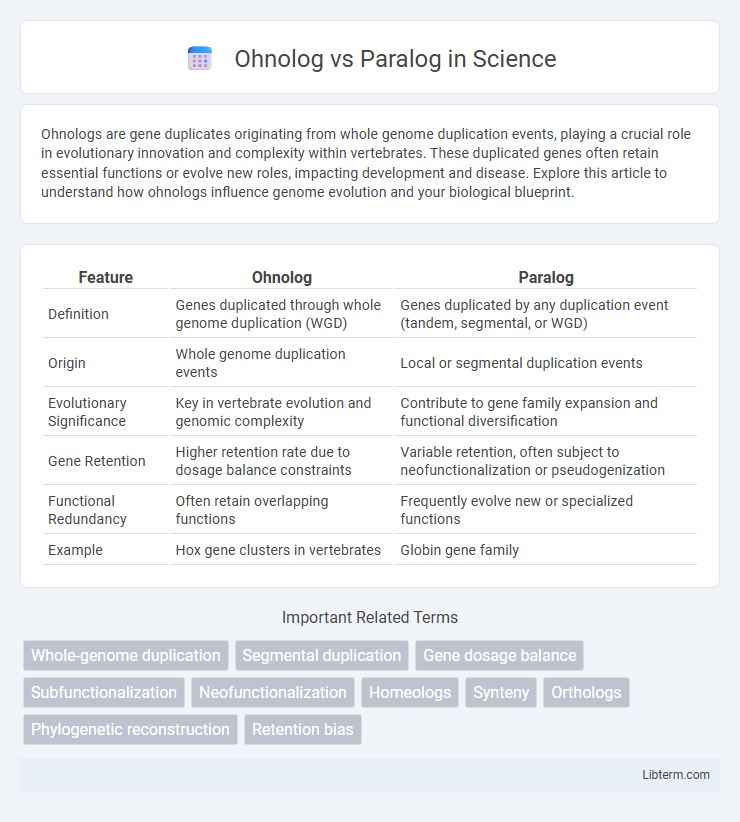
| Feature | Ohnolog | Paralog |
|---|---|---|
| Definition | Genes duplicated through whole genome duplication (WGD) | Genes duplicated by any duplication event (tandem, segmental, or WGD) |
| Origin | Whole genome duplication events | Local or segmental duplication events |
| Evolutionary Significance | Key in vertebrate evolution and genomic complexity | Contribute to gene family expansion and functional diversification |
| Gene Retention | Higher retention rate due to dosage balance constraints | Variable retention, often subject to neofunctionalization or pseudogenization |
| Functional Redundancy | Often retain overlapping functions | Frequently evolve new or specialized functions |
| Example | Hox gene clusters in vertebrates | Globin gene family |
Introduction to Ohnologs and Paralogs
Ohnologs are gene duplicates originating specifically from whole-genome duplication (WGD) events, playing a crucial role in evolutionary innovation and species diversification. Paralogs arise from any gene duplication event and can evolve new functions or subfunctions within a genome. Distinguishing ohnologs from paralogs provides insights into genomic architecture and the evolutionary history of gene families.
Definitions: Ohnologs vs Paralogs
Ohnologs are gene duplicates that arise specifically from whole genome duplication events, preserving large-scale genomic structures and often maintaining essential biological functions. Paralogs are genes related by duplication within a genome but can result from various duplication mechanisms including segmental duplication, and they may evolve new functions or specialize. Understanding the distinction between ohnologs and paralogs is crucial in evolutionary biology and genomics for studying gene function divergence and genome evolution.
Evolutionary Origins of Ohnologs
Ohnologs are gene duplicates that arise specifically from whole-genome duplication (WGD) events, distinguishing them from paralogs, which result from any gene duplication event including small-scale duplications. These ohnologs retain syntenic relationships and are often preserved due to dosage balance constraints influencing evolutionary trajectories. The evolutionary origins of ohnologs trace back to ancient WGD events in vertebrates, particularly notable in the two rounds of genome duplication known as the 2R hypothesis.
Evolutionary Origins of Paralogs
Paralogs originate from gene duplication events within a genome, resulting in multiple gene copies that evolve new functions over time. Ohnologs are a specific subset of paralogs arising from whole-genome duplication events, especially prevalent in vertebrate evolution. Understanding the evolutionary origins of paralogs, including ohnologs, is crucial for studying gene function diversification and genome complexity.
Mechanisms of Gene Duplication
Ohnologs arise from whole-genome duplications (WGDs), where entire sets of chromosomes are duplicated, leading to large-scale gene retention and functional diversification. Paralogs result from small-scale gene duplication events such as tandem duplications, segmental duplications, or replication slippage, generating gene copies within localized genomic regions. The distinct mechanisms underlying ohnolog and paralog formation influence the evolutionary trajectory and dosage sensitivity of duplicated genes.
Functional Divergence in Ohnologs and Paralogs
Ohnologs and paralogs both arise from gene duplication events but differ in their evolutionary origins, with ohnologs originating from whole-genome duplications and paralogs from local duplications. Functional divergence in ohnologs often reflects retained essential biological roles due to dosage balance constraints, leading to subfunctionalization or neofunctionalization with less redundancy than paralogs. Paralogs frequently exhibit greater functional divergence, driven by relaxed selective pressures allowing one copy to acquire novel functions while the other maintains the original gene role.
Genomic Distribution and Conservation
Ohnologs are gene duplicates that arise from whole-genome duplication events, resulting in their distribution across entire chromosomes or genomes, whereas paralogs emerge from smaller-scale gene duplication and tend to cluster in localized genomic regions. Ohnologs exhibit higher evolutionary conservation due to their essential roles in dosage balance and developmental processes, while paralogs often display more functional divergence and variability in sequence conservation. Genomic analyses reveal that ohnologs are preferentially retained in vertebrate genomes, highlighting their significance in maintaining genome stability and complex regulatory networks.
Biological Significance and Disease Associations
Ohnologs, originating from whole-genome duplications, play a crucial role in the evolution of complex gene networks and developmental processes, showing a strong enrichment in dosage-sensitive genes linked to developmental disorders and cancer. Paralogs, derived from smaller-scale gene duplication events, contribute to functional diversification and redundancy, often associated with adaptation and resilience but can also lead to genetic diseases when mutations affect only one copy. Both ohnologs and paralogs influence genetic robustness and pathogenesis, with ohnologs particularly implicated in dosage imbalance diseases and paralogs in subfunctionalization-related disorders.
Methods for Identifying Ohnologs and Paralogs
Ohnologs are duplicated genes originating from whole genome duplication events, identified primarily through synteny analysis and phylogenetic tree reconstruction to distinguish these from paralogs, which arise from smaller-scale duplication events such as tandem or segmental duplications. Methods like comparative genomics and molecular clock analysis help verify the timing of duplication events, enhancing the accuracy of distinguishing ohnologs from paralogs. Advanced bioinformatics tools, including gene collinearity mapping and sequence homology searches, are essential for detecting ohnologs by confirming their retention across large genomic blocks, unlike paralogs that often exhibit more localized duplication patterns.
Conclusion: Key Differences and Research Directions
Ohnologs arise from whole genome duplication events, whereas paralogs result from smaller-scale gene duplications, leading to distinct evolutionary trajectories. Ohnologs often retain crucial regulatory functions and exhibit dosage sensitivity, making them important for studying complex traits and disease associations. Future research should explore the mechanisms driving functional divergence between ohnologs and paralogs, with a focus on their roles in evolutionary innovation and genomic stability.
Ohnolog Infographic
 libterm.com
libterm.com