An operon is a cluster of genes regulated together, enabling efficient gene expression in prokaryotes by controlling transcription as a single unit. This mechanism ensures that related proteins are produced simultaneously, responding swiftly to environmental changes to optimize cellular function. Discover how understanding operons can enhance your knowledge of genetic regulation throughout this article.
Table of Comparison
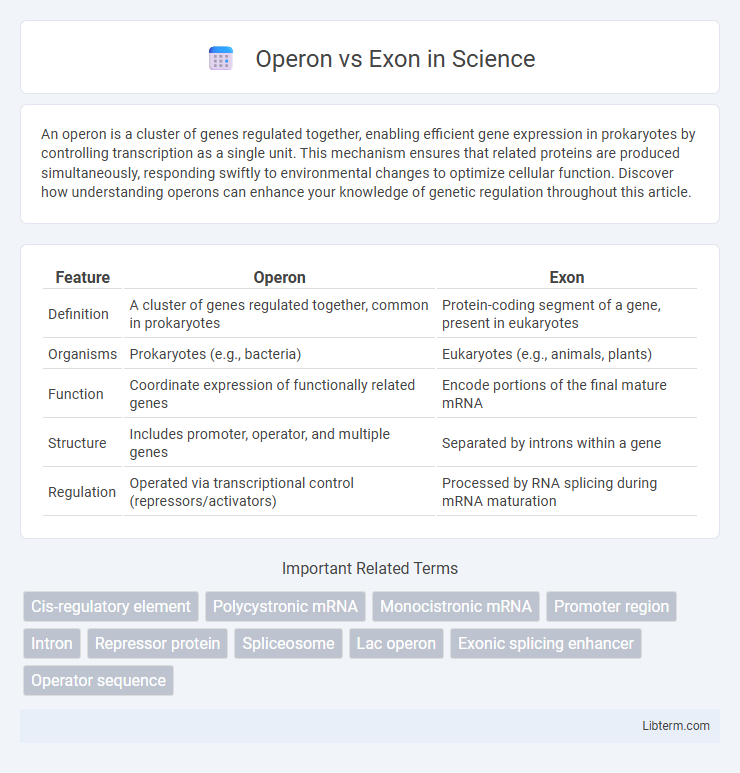
| Feature | Operon | Exon |
|---|---|---|
| Definition | A cluster of genes regulated together, common in prokaryotes | Protein-coding segment of a gene, present in eukaryotes |
| Organisms | Prokaryotes (e.g., bacteria) | Eukaryotes (e.g., animals, plants) |
| Function | Coordinate expression of functionally related genes | Encode portions of the final mature mRNA |
| Structure | Includes promoter, operator, and multiple genes | Separated by introns within a gene |
| Regulation | Operated via transcriptional control (repressors/activators) | Processed by RNA splicing during mRNA maturation |
Overview of Genetic Regulation: Operons and Exons
Operons are clusters of genes regulated together by a single promoter and operator, primarily found in prokaryotes, enabling coordinated expression of functionally related genes like the lac operon in E. coli. Exons, conversely, are coding sequences within eukaryotic genes spliced together during mRNA processing to form the final transcript, with intervening introns removed. Operons regulate gene transcription collectively at the DNA level, while exons contribute to gene expression diversity through alternative splicing at the RNA level.
Definitions: What Are Operons and Exons?
Operons are functional units of DNA in prokaryotes consisting of a cluster of genes regulated together under a single promoter, enabling coordinated gene expression. Exons are sequences within eukaryotic genes that code for portions of the final mature mRNA and are retained after RNA splicing. Operons control gene regulation in bacteria, while exons represent the coding regions in eukaryotic gene architecture.
Distinguishing Features: Operon Structure vs. Exon Structure
Operons consist of a cluster of genes regulated together under a single promoter, primarily found in prokaryotes, allowing coordinated transcription into a single mRNA transcript. Exons, in eukaryotic genomes, are coding regions within a gene separated by introns, and are spliced together during RNA processing to form a mature mRNA. The operon structure enables simultaneous expression of multiple genes, whereas exon structure facilitates alternative splicing, producing diverse protein isoforms from one gene.
Operons in Prokaryotic Gene Expression
Operons are clusters of genes in prokaryotic DNA that are transcribed together under the control of a single promoter, enabling coordinated gene expression. This organization allows bacteria to efficiently regulate metabolic pathways by turning multiple genes on or off simultaneously in response to environmental changes. Unlike exons, which are coding sequences within eukaryotic genes, operons function as a fundamental regulatory unit in prokaryotic gene expression.
Exons in Eukaryotic Gene Expression
Exons are the coding sequences in eukaryotic genes that remain in the mature mRNA after RNA splicing, playing a critical role in defining the amino acid sequence of the resulting protein. Unlike operons found in prokaryotes, which cluster multiple genes under a single promoter for coordinated expression, eukaryotic exons are interspersed with non-coding introns and allow for alternative splicing mechanisms that enhance protein diversity. The precise removal of introns and joining of exons is essential for accurate gene expression and functional protein synthesis in eukaryotic cells.
Transcriptional Control: Operons vs. Exons
Operons are clusters of genes regulated together by a single promoter, enabling coordinated transcriptional control in prokaryotes, whereas exons are coding sequences within genes in eukaryotes that are spliced during mRNA processing. Transcriptional control in operons involves regulatory proteins binding to operator sites to modulate the transcription of multiple genes simultaneously. In contrast, transcriptional control in eukaryotes includes the selective inclusion of exons through alternative splicing, affecting the diversity of the mRNA and protein products.
Functional Roles in Cellular Processes
Operons are clusters of genes regulated together to coordinate the production of proteins involved in related cellular functions, primarily found in prokaryotes, enabling efficient control of metabolic pathways. Exons are coding sequences within eukaryotic genes that are spliced together during mRNA processing to produce functional proteins essential for diverse cellular activities. While operons manage gene expression at the transcriptional level, exons contribute to protein diversity and function through post-transcriptional modification.
Evolutionary Significance of Operons and Exons
Operons, primarily found in prokaryotes, enable coordinated regulation of genes with related functions, promoting efficient gene expression and metabolic control, which enhances adaptability and survival in fluctuating environments. Exons, characteristic of eukaryotic genomes, facilitate complex gene structures allowing alternative splicing, leading to protein diversity and evolutionary flexibility in multicellular organisms. The evolutionary significance lies in operons providing streamlined, collective gene regulation for rapid response, while exons contribute to functional complexity and innovation through modular gene assembly.
Key Differences in Genetic Organization
Operons are clusters of genes transcribed as a single mRNA unit in prokaryotes, allowing coordinated regulation of genes with related functions, whereas exons are coding sequences within a single gene in eukaryotes that are spliced together to form mature mRNA. Operons include promoters, operators, and structural genes, enabling simultaneous expression control, while exons are separated by introns and processed post-transcriptionally. The key difference lies in operons functioning as gene group regulatory units in prokaryotes, whereas exons represent discrete coding segments within individual genes in eukaryotes.
Comparative Summary: Operons vs. Exons
Operons are clusters of genes regulated together, primarily found in prokaryotes, enabling coordinated expression of multiple proteins from a single mRNA transcript. Exons are coding sequences within eukaryotic genes that remain in the mature mRNA after splicing, responsible for encoding protein domains. Unlike operons, exons do not function collectively for gene regulation but are integral parts of individual gene transcripts contributing to protein diversity through alternative splicing.
Operon Infographic
 libterm.com
libterm.com