An operator is a symbol or function that performs specific mathematical, logical, or computational tasks on one or more operands. Understanding different types of operators, such as arithmetic, relational, and logical, is essential for efficient programming and problem-solving. Explore the rest of the article to deepen your knowledge of how operators can optimize your coding skills.
Table of Comparison
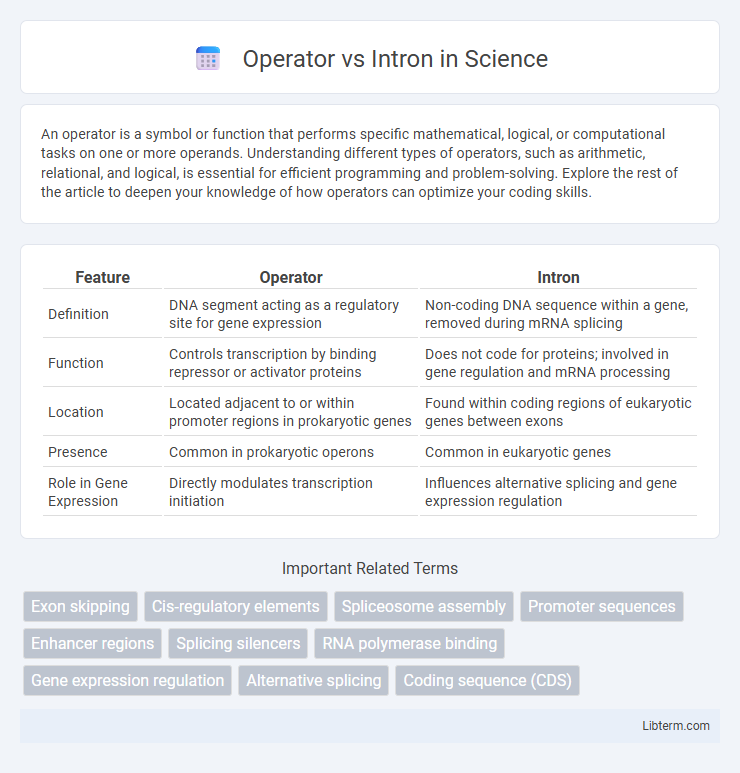
| Feature | Operator | Intron |
|---|---|---|
| Definition | DNA segment acting as a regulatory site for gene expression | Non-coding DNA sequence within a gene, removed during mRNA splicing |
| Function | Controls transcription by binding repressor or activator proteins | Does not code for proteins; involved in gene regulation and mRNA processing |
| Location | Located adjacent to or within promoter regions in prokaryotic genes | Found within coding regions of eukaryotic genes between exons |
| Presence | Common in prokaryotic operons | Common in eukaryotic genes |
| Role in Gene Expression | Directly modulates transcription initiation | Influences alternative splicing and gene expression regulation |
Introduction to Operators and Introns
Operators are DNA sequences that act as regulatory elements controlling gene expression by binding repressor proteins and thereby inhibiting transcription. Introns are non-coding regions within a gene that are removed during RNA splicing to produce mature messenger RNA. Understanding operators and introns is essential for studying gene regulation and the post-transcriptional modification processes in molecular biology.
Defining Operators in Genetic Regulation
Operators are specific DNA sequences located near promoters that act as binding sites for regulatory proteins, controlling gene expression by influencing RNA polymerase activity. In genetic regulation, operators function as checkpoints where repressors or activators bind to either inhibit or promote transcription, thereby modulating gene activity. Unlike introns, which are non-coding sequences removed during mRNA processing, operators directly impact transcription initiation and are key elements in operon systems such as the lac operon in prokaryotes.
Understanding Introns in Gene Structure
Introns are non-coding sequences within a gene that are transcribed into RNA but removed during RNA splicing, distinguishing them from coding exons. Operators serve as regulatory DNA elements that control gene expression by binding repressors or activators, typically located near or within operons in prokaryotes. Understanding introns in gene structure reveals their crucial role in alternative splicing, enabling the generation of multiple protein isoforms and increasing functional diversity.
Functional Roles: Operator vs Intron
Operators act as key regulatory DNA sequences where repressor proteins bind to control the transcription of adjacent genes, thus playing a crucial role in gene expression modulation. Introns, found within the coding regions of eukaryotic genes, are non-coding sequences removed during RNA splicing, influencing mRNA maturation and enabling alternative splicing for protein diversity. The functional distinction lies in operators governing transcription initiation, while introns primarily affect post-transcriptional processing and gene expression complexity.
Evolutionary Significance of Operators and Introns
Operators and introns play distinct roles in gene regulation and evolution, with operators serving as key regulatory DNA sequences that control gene expression in prokaryotes, enabling rapid adaptive responses to environmental changes. Introns, non-coding sequences within genes of eukaryotes, facilitate genetic diversity through alternative splicing and exon shuffling, driving evolutionary innovation and complexity. The evolutionary significance of operators lies in optimizing gene regulation efficiency, while introns contribute to functional diversification and the evolution of complex organisms.
Impact on Gene Expression: Operators and Introns
Operators regulate gene expression by acting as binding sites for repressor proteins that control the transcription of adjacent genes, thus directly influencing the activation or repression of gene clusters often seen in prokaryotes. Introns, found within eukaryotic genes, impact gene expression through alternative splicing mechanisms that generate multiple mRNA variants from a single gene, enhancing protein diversity and regulation complexity. Both elements play crucial roles in gene expression modulation but operate at different genetic levels and mechanisms--operators control transcription initiation, whereas introns regulate post-transcriptional mRNA processing.
Molecular Mechanisms: Operator Binding vs Intron Splicing
The operator functions as a DNA regulatory sequence where repressor proteins bind to control gene transcription by preventing RNA polymerase attachment, ensuring precise gene expression regulation. Introns are non-coding RNA segments removed from pre-mRNA through complex spliceosome-mediated splicing, involving small nuclear ribonucleoproteins (snRNPs) that recognize specific splice sites. Operator binding influences transcription initiation at the DNA level, whereas intron splicing modifies RNA transcripts post-transcriptionally to produce mature mRNA.
Examples in Prokaryotic and Eukaryotic Systems
Operators in prokaryotic systems, such as the lac operon in *Escherichia coli*, function as DNA sequences where repressors bind to regulate gene expression by controlling RNA polymerase access. In contrast, introns in eukaryotic organisms like humans are non-coding sequences within genes that are removed during RNA splicing to produce mature mRNA. The operator's regulatory role exemplifies transcriptional control in prokaryotes, whereas introns highlight post-transcriptional modifications fundamental to eukaryotic gene expression.
Biotechnological Applications Involving Operators and Introns
Operators serve as regulatory DNA sequences that control gene expression by binding repressor proteins, enabling precise modulation of metabolic pathways in engineered microorganisms; this is critical for optimizing synthetic biology circuits and producing recombinant proteins efficiently. Introns, non-coding sequences within genes, play a pivotal role in alternative splicing, allowing the generation of multiple protein isoforms, which enhances the versatility of gene expression in biotechnological applications such as gene therapy and molecular cloning. The strategic manipulation of operators and introns facilitates refined control over gene expression and protein diversity, driving advancements in metabolic engineering, therapeutic development, and functional genomics.
Future Research Directions: Operator and Intron Studies
Future research on operators and introns is poised to unravel their roles in gene regulation and evolutionary adaptation through advanced genomic editing and high-throughput sequencing techniques. Investigations into operator sequences aim to enhance synthetic biology applications by improving gene expression control, while studies on intron splicing mechanisms promise advancements in RNA therapeutics and precision medicine. Integrative approaches combining bioinformatics and functional genomics will accelerate discoveries on chromatin dynamics and non-coding RNA functions linked to operators and introns.
Operator Infographic
 libterm.com
libterm.com