A paralog is a gene related to another gene within the same organism, arising through gene duplication and often evolving new functions. Understanding paralogs is essential for interpreting genetic diversity and functional innovation in genomes. Explore the rest of the article to discover how paralogs impact evolutionary biology and medical research.
Table of Comparison
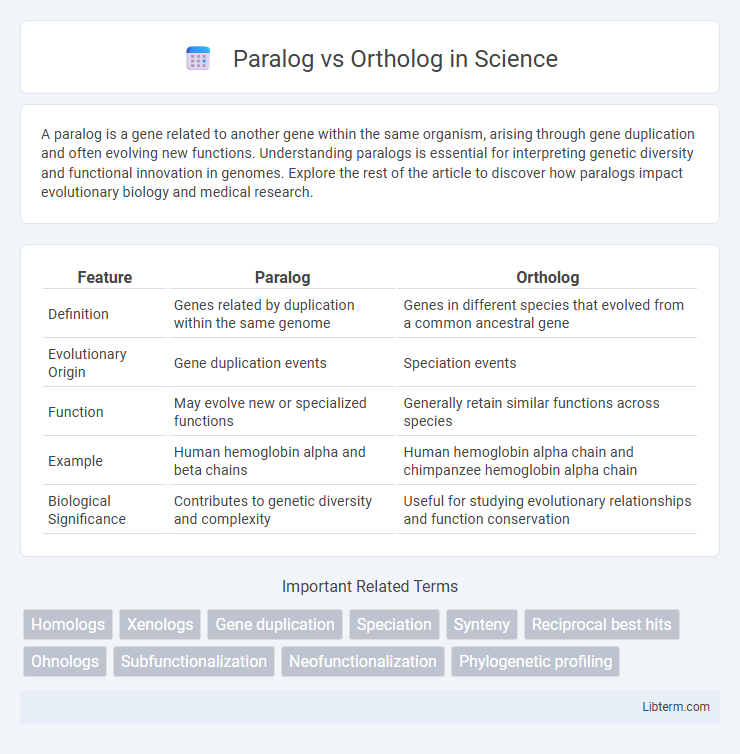
| Feature | Paralog | Ortholog |
|---|---|---|
| Definition | Genes related by duplication within the same genome | Genes in different species that evolved from a common ancestral gene |
| Evolutionary Origin | Gene duplication events | Speciation events |
| Function | May evolve new or specialized functions | Generally retain similar functions across species |
| Example | Human hemoglobin alpha and beta chains | Human hemoglobin alpha chain and chimpanzee hemoglobin alpha chain |
| Biological Significance | Contributes to genetic diversity and complexity | Useful for studying evolutionary relationships and function conservation |
Introduction to Paralog and Ortholog
Paralogs are genes within the same species that arise from gene duplication events and often evolve new functions, whereas orthologs are genes in different species that originated from a common ancestral gene through speciation and typically retain the same function. Identifying paralogous and orthologous relationships is crucial in comparative genomics to understand gene evolution and functional conservation. Methods such as sequence alignment, phylogenetic analysis, and synteny mapping help distinguish paralogs from orthologs by tracing evolutionary lineage and gene divergence.
Definitions of Paralog and Ortholog Genes
Paralog genes are homologous sequences that arise from gene duplication within the same species, often evolving new functions while retaining structural similarities. Ortholog genes, in contrast, are homologous genes separated by a speciation event, typically maintaining the same function across different species. Understanding the distinction between paralogs and orthologs is crucial for studying gene evolution, functional annotation, and comparative genomics.
Evolutionary Origins and Divergence
Paralogs are genes that arise from gene duplication events within the same genome, leading to gene copies that may evolve new functions over time. Orthologs, on the other hand, originate from a speciation event where a gene in an ancestral species diverges into two separate species, maintaining similar functions. Evolutionary divergence in paralogs often results in functional innovation, while orthologs generally retain conserved roles across different species.
Mechanisms of Gene Duplication and Speciation
Paralogs arise through gene duplication events within a genome, leading to multiple copies of a gene that may evolve new functions. Orthologs result from speciation, where a single gene in an ancestral species diverges into corresponding genes in descendant species. Gene duplication mechanisms include unequal crossing over, retrotransposition, and whole-genome duplication, while speciation involves reproductive isolation and genetic divergence.
Functional Divergence: Paralog vs Ortholog
Paralogs arise from gene duplication events and often evolve new or specialized functions, leading to functional divergence within the same organism. Orthologs originate from speciation events and typically retain similar functions across different species, maintaining functional conservation. Functional divergence between paralogs enables adaptation and complexity, whereas orthologs provide essential comparative insights into gene function and evolution.
Methods for Identifying Paralog and Ortholog Genes
Methods for identifying paralog and ortholog genes rely heavily on comparative genomics and phylogenetic analysis. Paralog genes are typically detected through gene duplication events using sequence similarity searches like BLAST combined with clustering algorithms such as OrthoMCL or InParanoid. Orthologs are identified by constructing phylogenetic trees and establishing reciprocal best hits (RBH) between species, supplemented by synteny analysis to confirm evolutionary relationships.
Importance in Comparative Genomics
Paralog and ortholog genes play crucial roles in comparative genomics by providing insights into gene function and evolutionary relationships. Orthologs, arising from speciation events, help identify conserved functions across species, while paralogs, resulting from gene duplication within a genome, reveal functional diversification and innovation. Distinguishing between paralogous and orthologous genes is essential for accurate gene annotation, evolutionary studies, and understanding organismal adaptation.
Implications for Phylogenetic Analysis
Paralogs and orthologs represent different evolutionary relationships, where orthologs arise from speciation events and retain similar functions across species, while paralogs result from gene duplication and may evolve new functions. Accurate identification of orthologous genes is crucial for reliable phylogenetic analysis, as it ensures the inferred evolutionary relationships reflect species divergence rather than gene duplication events. Misinterpreting paralogs as orthologs can lead to erroneous phylogenetic trees and misestimation of divergence times.
Role in Disease and Functional Annotation
Paralogs often contribute to genetic redundancy, complicating disease gene identification due to their overlapping functions, while orthologs provide clearer insights into conserved disease mechanisms across species. Functional annotation of paralogs requires careful differentiation to avoid misinterpretation of gene function, whereas ortholog annotation leverages evolutionary conservation to predict gene roles more reliably. Distinguishing between paralogous and orthologous genes enhances the accuracy of disease gene prioritization and improves the transferability of functional annotations in comparative genomics.
Summary and Future Perspectives
Paralogs are genes related by duplication within a genome, often evolving new functions, while orthologs are genes in different species that originated from a common ancestral gene and usually retain the same function. Understanding the divergence and conservation patterns between paralogs and orthologs improves functional annotation and evolutionary studies across diverse organisms. Future research integrating high-throughput genomics and advanced phylogenetics promises to refine gene function prediction and clarify evolutionary relationships at greater resolution.
Paralog Infographic
 libterm.com
libterm.com