Numerical taxonomy uses quantitative methods to classify organisms based on measurable characteristics, ensuring objective and reproducible categorization. By analyzing large data sets with statistical techniques, it reveals natural relationships among species that may be overlooked in traditional classification. Explore the rest of the article to understand how numerical taxonomy can enhance your approach to biological classification.
Table of Comparison
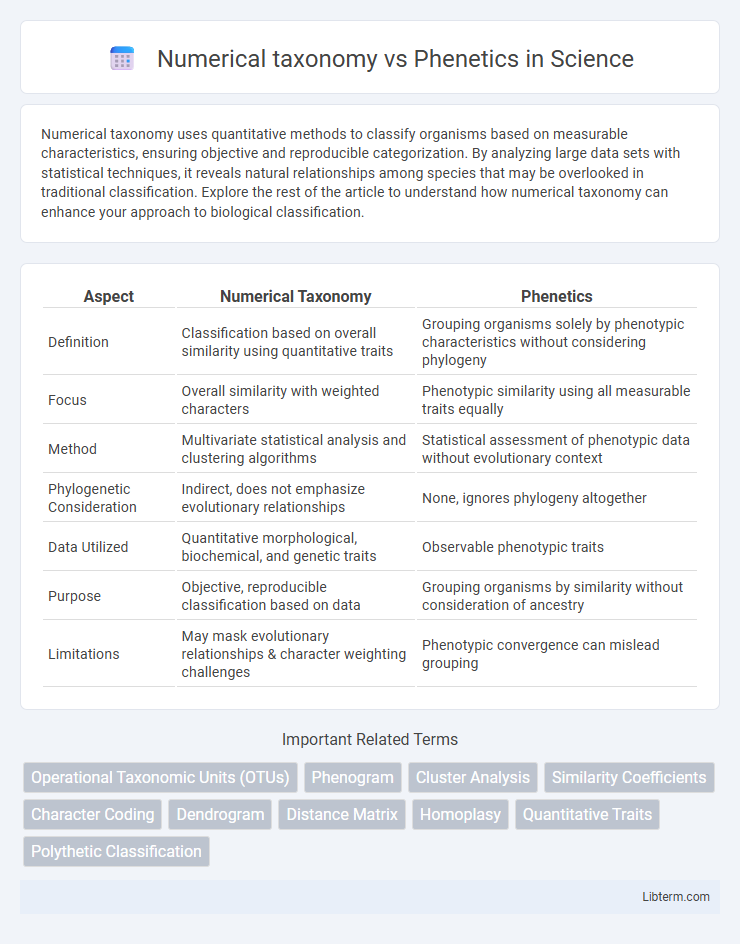
| Aspect | Numerical Taxonomy | Phenetics |
|---|---|---|
| Definition | Classification based on overall similarity using quantitative traits | Grouping organisms solely by phenotypic characteristics without considering phylogeny |
| Focus | Overall similarity with weighted characters | Phenotypic similarity using all measurable traits equally |
| Method | Multivariate statistical analysis and clustering algorithms | Statistical assessment of phenotypic data without evolutionary context |
| Phylogenetic Consideration | Indirect, does not emphasize evolutionary relationships | None, ignores phylogeny altogether |
| Data Utilized | Quantitative morphological, biochemical, and genetic traits | Observable phenotypic traits |
| Purpose | Objective, reproducible classification based on data | Grouping organisms by similarity without consideration of ancestry |
| Limitations | May mask evolutionary relationships & character weighting challenges | Phenotypic convergence can mislead grouping |
Introduction to Numerical Taxonomy and Phenetics
Numerical taxonomy is a quantitative approach to classifying organisms based on overall similarity using statistical methods and numerical algorithms to analyze multiple traits simultaneously. Phenetics, closely related to numerical taxonomy, emphasizes the classification of organisms purely by observable characteristics without considering evolutionary relationships, aiming to group species by overall phenotypic resemblance. Both approaches utilize mathematical tools to generate phenograms or dendrograms, facilitating objective assessments of biological similarity for taxonomy and systematics.
Historical Development of Both Approaches
Numerical taxonomy, established in the mid-20th century by Robert Sokal and Peter Sneath, introduced quantitative methods to classify organisms based on overall similarity using numerical data. Phenetics, developed concurrently, emphasized measuring overall phenotypic similarity without evolutionary considerations, relying mainly on observable traits and statistical analyses. Both approaches marked a shift from traditional taxonomy by prioritizing objective, data-driven classifications and influenced modern systematic biology by integrating computational methods into taxonomy.
Key Principles of Numerical Taxonomy
Numerical taxonomy relies on quantitative measurements and statistical analysis to classify organisms based on overall similarity, using a large set of characters weighted equally. It emphasizes objective, reproducible clustering methods such as phenograms constructed through algorithms like unweighted pair group method with arithmetic mean (UPGMA). This approach contrasts with traditional phenetics by prioritizing numerical data processing to reduce subjective bias in taxonomic classification.
Fundamental Concepts of Phenetics
Phenetics, also known as numerical taxonomy, emphasizes the classification of organisms based on overall similarity using quantitative measurements of multiple traits, without considering phylogenetic relationships. Its fundamental concept involves the use of numerical algorithms and statistical methods to cluster organisms into groups based on phenotype data, producing phenograms that reflect patterns of similarity. Phenetics relies heavily on metrics such as similarity coefficients and distance measures to generate objective, reproducible classifications grounded in observable characteristics.
Methodologies: Data Collection and Analysis
Numerical taxonomy employs quantitative methods for data collection, focusing on measurable traits such as morphological measurements and genetic markers, which are analyzed using multivariate statistical techniques like cluster analysis and principal component analysis. Phenetics, on the other hand, relies on overall similarity among organisms, often incorporating both qualitative and quantitative traits, and uses distance or similarity coefficients to group taxa based on phenotypic resemblance. Both methodologies emphasize objective classification, but numerical taxonomy prioritizes numerical algorithms for data processing, whereas phenetics primarily evaluates phenotypic patterns without explicit phylogenetic considerations.
Similarities Between Numerical Taxonomy and Phenetics
Numerical taxonomy and phenetics both rely on the quantitative analysis of multiple traits to classify organisms based on overall similarity without emphasizing evolutionary relationships. Both methods employ multivariate statistical techniques such as cluster analysis and principal component analysis to group taxa objectively. The primary goal in each approach is to provide standardized, reproducible classifications by minimizing subjective bias in trait selection and comparison.
Differences in Classification Approaches
Numerical taxonomy emphasizes quantitative analysis of multiple traits using statistical methods to create classifications based on overall similarity, while phenetics groups organisms purely on observable characteristics without considering evolutionary relationships. Numerical taxonomy employs algorithms and cluster analysis to generate objective classifications, whereas phenetics relies more on subjective assessment of phenotypic traits. Consequently, numerical taxonomy provides a more reproducible, data-driven framework, contrasting with phenetics' traditional, morphology-based grouping system.
Advantages and Limitations of Each Method
Numerical taxonomy leverages quantitative data and statistical techniques to classify organisms, offering precise and reproducible results, yet it may overlook ecological or evolutionary significance due to heavy reliance on measurable traits. Phenetics assesses overall similarity without distinguishing between ancestral and derived traits, providing a straightforward clustering approach but often leading to misleading classifications by not considering phylogenetic relationships. Both methods facilitate systematic grouping, but numerical taxonomy emphasizes objectivity and reproducibility while phenetics offers simplicity at the cost of evolutionary accuracy.
Applications in Modern Biological Research
Numerical taxonomy applies quantitative methods to classify organisms based on measurable traits, enabling precise phylogenetic analysis and biodiversity assessment. Phenetics groups organisms by overall similarity using statistical techniques, facilitating rapid identification and environmental monitoring. Both approaches support modern biological research by improving species delimitation, ecological studies, and evolutionary pattern recognition through advanced computational tools.
Future Trends and Perspectives in Taxonomy
Numerical taxonomy and phenetics are increasingly integrated with molecular techniques to enhance species classification accuracy and evolutionary insights, leveraging large-scale genomic data and machine learning algorithms. Future trends emphasize automating data analysis and developing dynamic, interactive taxonomic databases to improve reproducibility and international collaboration. Advances in bioinformatics and high-throughput sequencing tools will drive precision taxonomy, enabling real-time monitoring of biodiversity and adaptive changes across ecosystems.
Numerical taxonomy Infographic
 libterm.com
libterm.com