Introns are non-coding segments of DNA within a gene that are transcribed into RNA but removed during mRNA processing before translation. Understanding their role is crucial for grasping genetic regulation and expression mechanisms. Explore the rest of this article to learn how introns influence gene function and impact your genetic makeup.
Table of Comparison
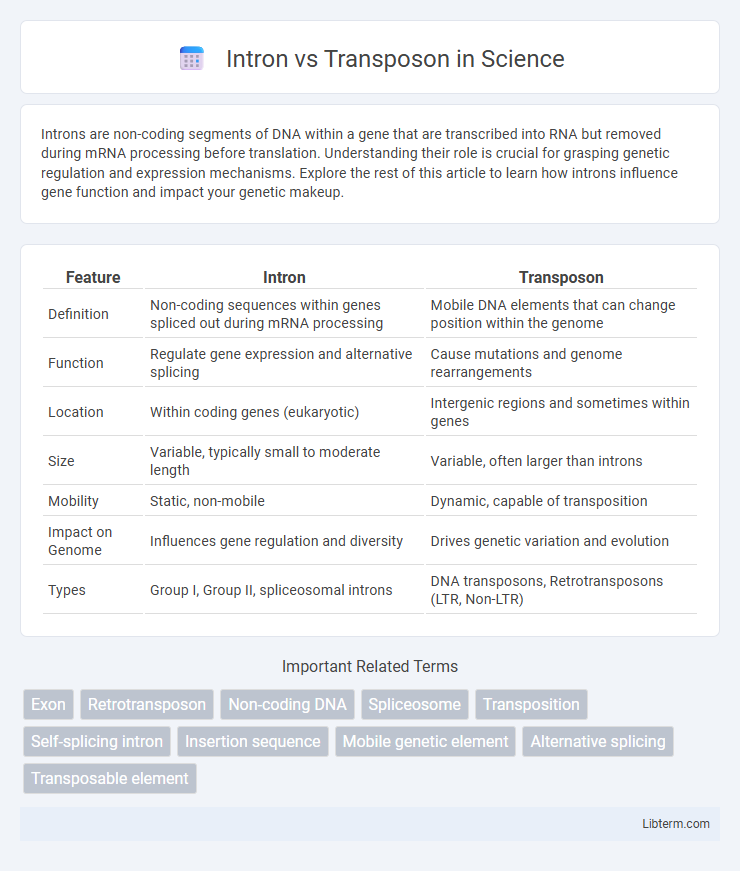
| Feature | Intron | Transposon |
|---|---|---|
| Definition | Non-coding sequences within genes spliced out during mRNA processing | Mobile DNA elements that can change position within the genome |
| Function | Regulate gene expression and alternative splicing | Cause mutations and genome rearrangements |
| Location | Within coding genes (eukaryotic) | Intergenic regions and sometimes within genes |
| Size | Variable, typically small to moderate length | Variable, often larger than introns |
| Mobility | Static, non-mobile | Dynamic, capable of transposition |
| Impact on Genome | Influences gene regulation and diversity | Drives genetic variation and evolution |
| Types | Group I, Group II, spliceosomal introns | DNA transposons, Retrotransposons (LTR, Non-LTR) |
Introduction to Introns and Transposons
Introns are non-coding sequences within genes that are removed during RNA splicing, playing a crucial role in gene expression regulation and alternative splicing. Transposons, also known as "jumping genes," are mobile genetic elements that can move within the genome, contributing to genetic variation and genome evolution. Both introns and transposons influence genome architecture but differ in function and mobility.
Defining Introns: Structure and Function
Introns are non-coding sequences within genes that are transcribed into RNA but removed during RNA splicing to produce mature mRNA. They contribute to gene regulation, alternative splicing, and genomic diversity by allowing for multiple protein isoforms. Unlike transposons, which are mobile genetic elements that can move around the genome, introns remain fixed within a gene and primarily influence gene expression and RNA processing.
Understanding Transposons: Types and Mechanisms
Transposons, also known as "jumping genes," are DNA sequences that can change their position within the genome, affecting gene function and genomic stability. They are classified primarily into two types: Class I transposons (retrotransposons), which move via an RNA intermediate using a "copy-and-paste" mechanism, and Class II transposons (DNA transposons), which mobilize directly through a "cut-and-paste" mechanism. Understanding the mechanisms of transposons involves studying key enzymes like transposase for DNA transposons and reverse transcriptase for retrotransposons, which facilitate their excision, replication, and reintegration into new genomic locations.
Genomic Distribution: Introns vs Transposons
Introns are non-coding sequences found within the genes of eukaryotic organisms, interrupting the coding regions (exons) and predominantly distributed throughout the nuclear genome. Transposons, or transposable elements, are mobile genetic sequences scattered extensively across both prokaryotic and eukaryotic genomes, often constituting a significant portion of genomic DNA, such as approximately 45% of the human genome. While introns are typically localized within gene boundaries, transposons can be found in intergenic regions as well as within genes, contributing to genomic diversity and structural variation.
Evolutionary Significance of Introns and Transposons
Introns contribute to genetic diversity and evolution by enabling alternative splicing, which increases protein variability without expanding genome size. Transposons act as mobile genetic elements that drive genome rearrangement, gene duplication, and horizontal gene transfer, profoundly influencing genome evolution and adaptation. Both elements shape genomic architecture, with introns enhancing regulatory complexity and transposons promoting genomic innovation through their mobility.
Impact on Gene Expression and Regulation
Introns play a critical role in gene expression by enabling alternative splicing, allowing a single gene to produce multiple protein variants and increasing proteomic diversity. Transposons impact gene regulation by inserting themselves into or near genes, often disrupting normal gene function or altering regulatory regions, which can lead to gene silencing or activation through epigenetic modifications. Both introns and transposons contribute to genome evolution and complexity by modulating transcriptional and post-transcriptional processes.
Role in Genome Evolution and Plasticity
Introns contribute to genome evolution by facilitating alternative splicing, which increases proteomic diversity without altering the DNA sequence. Transposons drive genome plasticity through their ability to move within the genome, causing mutations, gene duplications, and chromosomal rearrangements that create genetic variability. Both elements play critical roles in shaping genome architecture and enabling adaptive evolutionary responses.
Implications in Genetic Disorders and Diseases
Introns, non-coding regions within genes, influence genetic disorders by affecting RNA splicing, leading to abnormal protein production and diseases like spinal muscular atrophy. Transposons, mobile genetic elements, contribute to genomic instability by inserting themselves into crucial genes, causing mutations linked to conditions such as hemophilia and cancer. Understanding the distinct roles of introns and transposons in gene expression and genome dynamics is critical for developing targeted therapies for genetic disorders.
Molecular Techniques for Studying Introns and Transposons
Molecular techniques such as PCR amplification and DNA sequencing are pivotal for analyzing intron-exon boundaries and transposon insertion sites, enabling precise characterization of their sequences and functions. RNA sequencing (RNA-seq) provides deep insights into intron splicing patterns and alternative splicing events, while transposon display and inverse PCR facilitate the detection and mapping of transposable elements within genomes. CRISPR-Cas9 genome editing further allows targeted manipulation of introns and transposons to study their roles in gene regulation and genome evolution.
Future Perspectives and Research Directions
Future research on introns and transposons is directed towards elucidating their roles in genome evolution, regulation, and adaptation. Advances in CRISPR-based technologies and long-read sequencing enable precise functional analysis and manipulation of intronic regions and transposable elements. Integrating multi-omics data will uncover their impact on gene expression dynamics, epigenetic modifications, and disease mechanisms, paving the way for novel therapeutic strategies.
Intron Infographic
 libterm.com
libterm.com