Numerical taxonomy uses quantitative methods to classify organisms based on measurable traits, enhancing the objectivity and reproducibility of species identification. This approach employs statistical techniques to analyze similarities and differences among biological specimens, facilitating more accurate and efficient classification. Discover how numerical taxonomy can transform your understanding of biodiversity by exploring the full article.
Table of Comparison
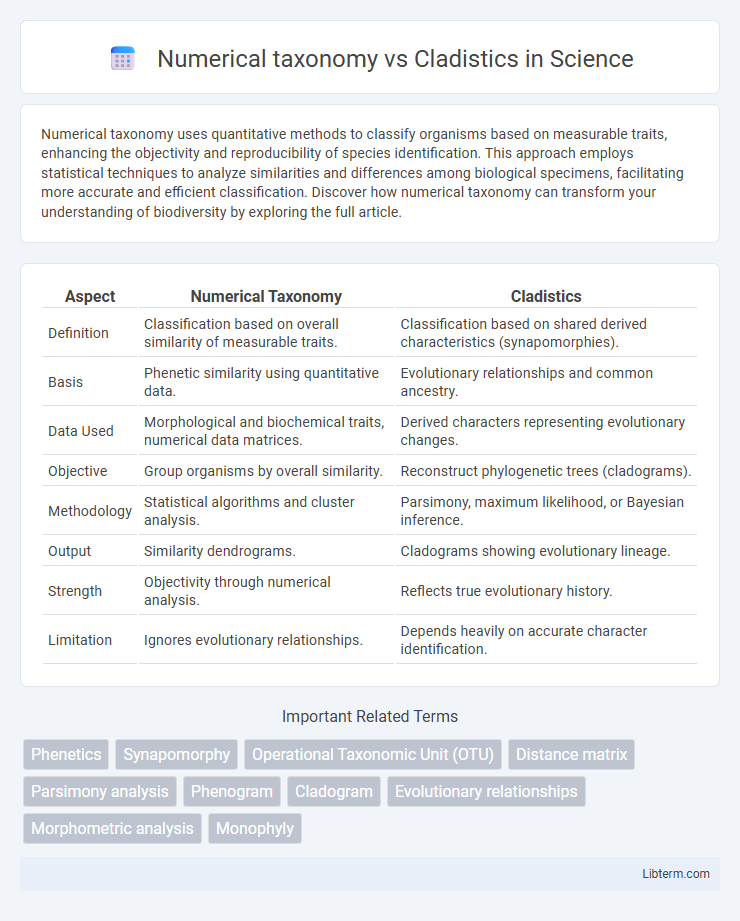
| Aspect | Numerical Taxonomy | Cladistics |
|---|---|---|
| Definition | Classification based on overall similarity of measurable traits. | Classification based on shared derived characteristics (synapomorphies). |
| Basis | Phenetic similarity using quantitative data. | Evolutionary relationships and common ancestry. |
| Data Used | Morphological and biochemical traits, numerical data matrices. | Derived characters representing evolutionary changes. |
| Objective | Group organisms by overall similarity. | Reconstruct phylogenetic trees (cladograms). |
| Methodology | Statistical algorithms and cluster analysis. | Parsimony, maximum likelihood, or Bayesian inference. |
| Output | Similarity dendrograms. | Cladograms showing evolutionary lineage. |
| Strength | Objectivity through numerical analysis. | Reflects true evolutionary history. |
| Limitation | Ignores evolutionary relationships. | Depends heavily on accurate character identification. |
Introduction to Numerical Taxonomy and Cladistics
Numerical taxonomy classifies organisms based on overall similarity using quantitative methods and phenotypic traits, employing algorithms like cluster analysis to generate classifications. Cladistics focuses on evolutionary relationships by analyzing shared derived characteristics (synapomorphies) to construct phylogenetic trees that reflect common ancestry. Both approaches utilize different principles: numerical taxonomy emphasizes measurable traits for grouping, while cladistics prioritizes lineage-based connections through cladograms.
Historical Development of Taxonomic Methods
Numerical taxonomy, developed in the mid-20th century by Robert Sokal and Peter Sneath, introduced a quantitative approach to classification using computer algorithms to assess overall similarity based on multiple traits. Cladistics, pioneered by Willi Hennig in the 1950s, shifted taxonomic focus toward evolutionary relationships by classifying organisms based on shared derived characteristics (synapomorphies) and common ancestry. The historical development of taxonomic methods reflects a transition from phenetic, similarity-based systems to phylogenetic, ancestry-based frameworks, profoundly influencing modern systematics and biodiversity studies.
Fundamental Principles of Numerical Taxonomy
Numerical taxonomy relies on quantitative methods to classify organisms based on overall similarity, using multivariate statistical techniques to analyze numerous traits simultaneously without emphasizing evolutionary relationships. Its fundamental principles include the use of phenetic data, objective measurements, and cluster analysis to group organisms into taxa that reflect observable characteristics rather than inferred ancestry. This approach contrasts with cladistics, which prioritizes shared derived characters and evolutionary lineage for phylogenetic classification.
Core Concepts of Cladistics
Cladistics centers on classifying organisms based on shared derived characteristics called synapomorphies, emphasizing evolutionary relationships and common ancestry. It constructs phylogenetic trees, or cladograms, to represent hypotheses of lineage divergence using monophyletic groups defined by unique innovations. Numerical taxonomy, by contrast, relies on overall similarity measures and phenetic clustering, often disregarding the evolutionary significance of traits.
Key Differences Between Numerical Taxonomy and Cladistics
Numerical taxonomy classifies organisms based on overall similarity using quantitative methods and phenotypic traits, while cladistics focuses on evolutionary relationships inferred from shared derived characteristics (synapomorphies). Numerical taxonomy employs cluster analysis and similarity coefficients without emphasizing phylogeny, whereas cladistics builds hierarchical trees (cladograms) that reflect common ancestry and lineage divergence. The primary distinction lies in numerical taxonomy's phenetic approach versus cladistics' phylogenetic framework for understanding biological classification.
Data Types and Analyses in Both Approaches
Numerical taxonomy primarily utilizes quantitative data, such as morphological measurements and biochemical traits, analyzed through cluster analysis and multivariate statistical techniques to group organisms based on overall similarity. Cladistics relies on qualitative, homologous character states derived from genetic or morphological data, using parsimony or maximum likelihood algorithms to reconstruct evolutionary relationships by identifying shared derived traits (synapomorphies). While numerical taxonomy emphasizes phenetic similarity without necessarily reflecting phylogeny, cladistics explicitly models evolutionary history and lineage divergence through character state transformations.
Strengths and Limitations of Numerical Taxonomy
Numerical taxonomy excels in objectively quantifying similarities among organisms using extensive data sets and statistical methods, which enhances reproducibility and minimizes subjective bias. Its reliance on overall phenotypic traits allows for comprehensive classification but may obscure evolutionary relationships by not distinguishing homologous from analogous characteristics. Limitations include sensitivity to the selection of characters and arbitrary weighting, which can affect the robustness and biological relevance of the resulting taxa.
Advantages and Challenges of Cladistics
Cladistics offers precise classification based on evolutionary relationships by analyzing shared derived characteristics, enhancing the accuracy of phylogenetic trees. Advantages include its ability to reveal true lineage connections and facilitate predictions about ancestral traits. Challenges involve the complexity of determining character states, potential homoplasy that can mislead analyses, and reliance on comprehensive and accurate morphological or genetic data.
Applications in Biological Classification
Numerical taxonomy applies quantitative methods to classify organisms based on overall similarity, utilizing statistical algorithms and multivariate analysis to generate phenetic trees that are widely used in microbial taxonomy and ecological studies. Cladistics emphasizes evolutionary relationships by analyzing shared derived characteristics (synapomorphies) to construct phylogenetic trees, crucial for understanding lineage divergence and evolutionary history in vertebrate and plant systematics. Both approaches complement each other in biological classification by combining phenetic data with phylogenetic inference to create robust, comprehensive taxonomies.
Future Perspectives in Systematic Methodologies
Future perspectives in systematic methodologies emphasize integrating numerical taxonomy's quantitative data analysis with cladistics' evolutionary relationships to enhance phylogenetic accuracy. Advances in computational algorithms and machine learning enable handling large genomic datasets, promoting more precise species classification and evolutionary inference. This hybrid approach fosters comprehensive biodiversity assessments and accelerates discoveries in molecular systematics and conservation biology.
Numerical taxonomy Infographic
 libterm.com
libterm.com