A regulatory element controls gene expression by interacting with specific proteins to modulate transcription levels, directly impacting cellular functions and organism development. These DNA sequences, including enhancers, silencers, and promoters, play a crucial role in gene activation or repression in response to environmental signals. Explore the rest of the article to understand how these elements influence your genetic landscape and biological outcomes.
Table of Comparison
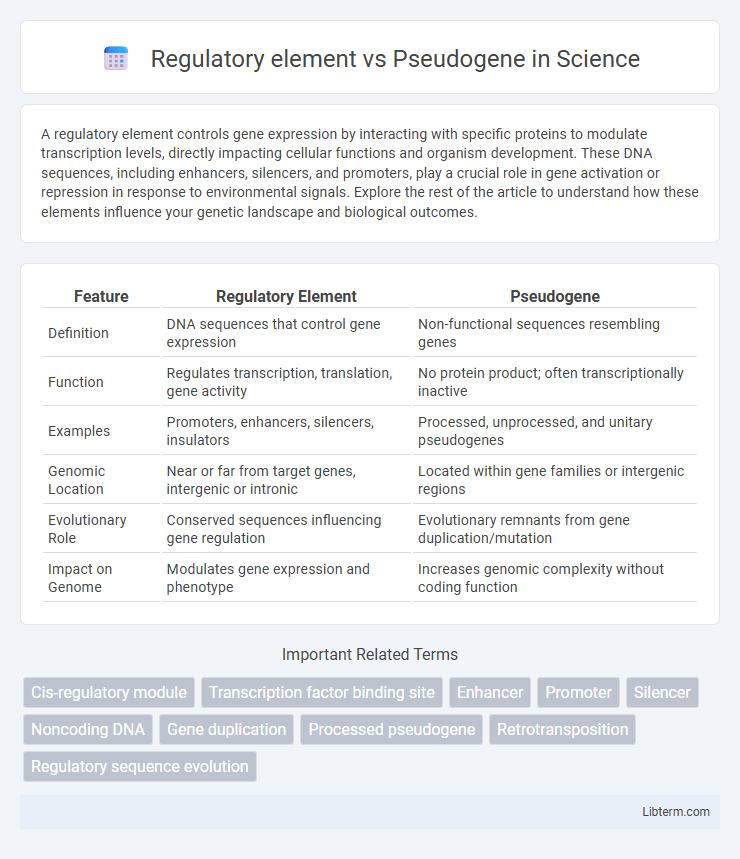
| Feature | Regulatory Element | Pseudogene |
|---|---|---|
| Definition | DNA sequences that control gene expression | Non-functional sequences resembling genes |
| Function | Regulates transcription, translation, gene activity | No protein product; often transcriptionally inactive |
| Examples | Promoters, enhancers, silencers, insulators | Processed, unprocessed, and unitary pseudogenes |
| Genomic Location | Near or far from target genes, intergenic or intronic | Located within gene families or intergenic regions |
| Evolutionary Role | Conserved sequences influencing gene regulation | Evolutionary remnants from gene duplication/mutation |
| Impact on Genome | Modulates gene expression and phenotype | Increases genomic complexity without coding function |
Introduction to Regulatory Elements and Pseudogenes
Regulatory elements are DNA sequences that control gene expression by acting as binding sites for transcription factors, influencing when, where, and how much a gene is expressed. Pseudogenes, on the other hand, are non-functional segments of DNA that resemble functional genes but contain mutations preventing their expression or protein production. Understanding the distinct roles of regulatory elements and pseudogenes is crucial for exploring gene regulation mechanisms and genome evolution.
Defining Regulatory Elements: Roles in Gene Expression
Regulatory elements are DNA sequences that control gene expression by interacting with transcription factors and other proteins, thereby influencing when, where, and how genes are activated or silenced. These elements include promoters, enhancers, silencers, and insulators, each playing critical roles in orchestrating precise gene expression patterns essential for cellular function and development. In contrast, pseudogenes are non-functional gene copies that do not directly regulate gene expression but may impact gene regulation indirectly through mechanisms like RNA interference or acting as genetic reservoirs.
Understanding Pseudogenes: Origins and Types
Pseudogenes originate from gene duplication or retrotransposition events, where functional genes accumulate mutations rendering them nonfunctional. They are classified into processed pseudogenes, lacking introns and derived from reverse-transcribed mRNA, and non-processed pseudogenes, which retain intron-exon structure but contain disabling mutations. Understanding these types provides insights into genome evolution and the regulatory potential of pseudogenes despite their non-coding status.
Structural Differences: Regulatory Elements vs. Pseudogenes
Regulatory elements are short DNA sequences such as promoters, enhancers, and silencers that control gene expression by binding transcription factors, whereas pseudogenes are non-functional DNA sequences resembling functional genes but harbor mutations preventing protein coding. Structurally, regulatory elements lack coding potential and often contain conserved motifs critical for transcriptional regulation, while pseudogenes possess gene-like sequences with disruptions like premature stop codons or frameshift mutations. The distinct sequence features and functional roles differentiate regulatory elements' compact, motif-rich regions from pseudogenes' gene-derived but non-expressed sequences.
Functional Significance in the Genome
Regulatory elements are critical DNA sequences that control gene expression by interacting with transcription factors and other proteins, ensuring precise spatial and temporal regulation of genes essential for development and cellular function. Pseudogenes, often considered non-functional remnants of genes, can influence genome regulation by acting as decoys for microRNAs or by producing regulatory RNAs that modulate gene expression. Understanding the functional significance of these genomic components reveals that while regulatory elements directly orchestrate gene activity, pseudogenes contribute indirectly to genome regulation through complex mechanisms impacting gene expression networks.
Evolutionary Perspectives: Regulatory Elements vs. Pseudogenes
Regulatory elements and pseudogenes represent distinct evolutionary mechanisms in genome function and adaptation. Regulatory elements, such as enhancers and silencers, have evolved to finely tune gene expression and facilitate phenotypic diversity, contributing to organismal complexity. In contrast, pseudogenes arise from gene duplication or retrotransposition events followed by loss of coding potential, serving as genomic fossils that may occasionally gain regulatory roles or influence gene expression through RNA-mediated mechanisms.
Identification Methods for Regulatory Elements and Pseudogenes
Identification methods for regulatory elements often involve experimental techniques such as chromatin immunoprecipitation sequencing (ChIP-seq) to detect DNA-binding proteins, and DNase I hypersensitivity assays to locate open chromatin regions indicative of active regulatory sites. Computational tools use sequence conservation, motif discovery, and machine learning models trained on epigenetic marks to predict regulatory elements genome-wide. Pseudogene identification relies on comparative genomics, recognizing sequences with high similarity to functional genes but containing disabling mutations like premature stop codons or frameshifts, often confirmed through RNA sequencing data showing lack of transcription or translational activity.
Case Studies: Impacts on Human Disease and Health
Regulatory elements, such as enhancers and promoters, control gene expression and have been linked to various human diseases by altering transcriptional activity, as demonstrated in case studies on cancer and autoimmune disorders. Pseudogenes, once considered non-functional, are increasingly recognized for their role in gene regulation and disease, influencing microRNA networks and acting as competitive endogenous RNAs in conditions like cancer and cardiovascular diseases. Recent research highlights that mutations or dysregulation in both regulatory elements and pseudogenes contribute significantly to the pathogenesis of complex diseases, offering novel insights for therapeutic interventions.
Regulatory Elements and Pseudogenes in Genomic Research
Regulatory elements are DNA sequences that control gene expression, playing a critical role in developmental processes and cellular function by binding transcription factors. Pseudogenes, once considered nonfunctional, are increasingly recognized for their potential regulatory roles, including acting as microRNA sponges or influencing gene expression through antisense transcripts. Genomic research leverages high-throughput sequencing and epigenomic mapping to elucidate the dynamic interaction between regulatory elements and pseudogenes, advancing the understanding of genetic regulation and disease mechanisms.
Future Directions and Challenges
Future directions in studying regulatory elements and pseudogenes involve advanced genomic editing technologies like CRISPR-Cas9 to elucidate their functional roles in gene expression and cellular processes. Challenges include distinguishing functional regulatory elements from non-functional sequences and understanding the diverse mechanisms by which pseudogenes influence gene regulation through RNA-based pathways or epigenetic modifications. Integrative multi-omics approaches and machine learning models hold promise for overcoming these challenges by improving the annotation and functional characterization of regulatory elements and pseudogenes in complex genomes.
Regulatory element Infographic
 libterm.com
libterm.com