Synthetic lethality occurs when the simultaneous impairment of two genes leads to cell death, while a defect in only one gene does not. This concept is pivotal in cancer therapy, enabling targeted treatments that kill cancer cells by exploiting specific genetic vulnerabilities. Explore the rest of the article to understand how synthetic lethality can transform personalized medicine.
Table of Comparison
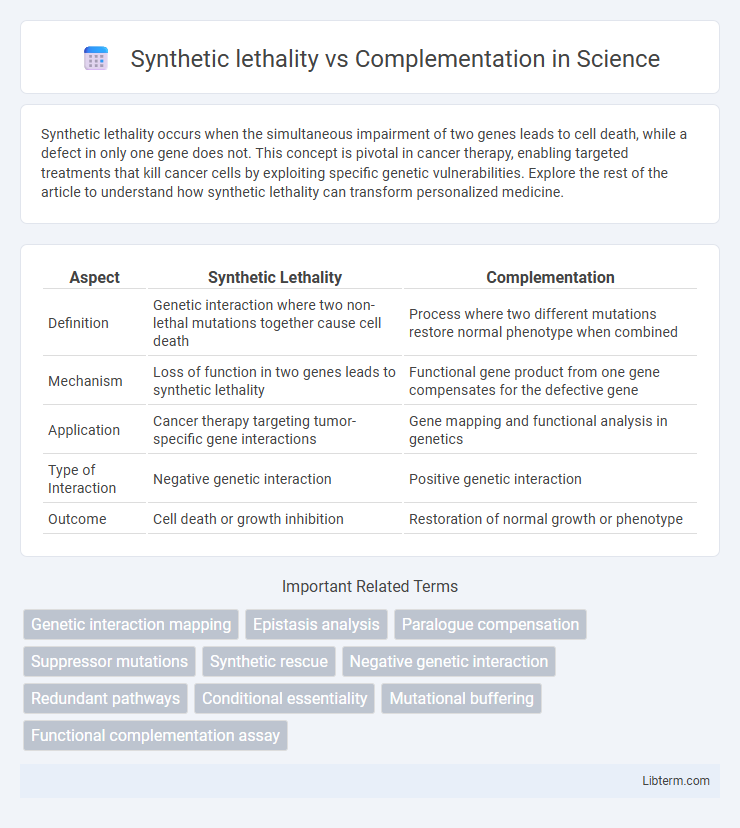
| Aspect | Synthetic Lethality | Complementation |
|---|---|---|
| Definition | Genetic interaction where two non-lethal mutations together cause cell death | Process where two different mutations restore normal phenotype when combined |
| Mechanism | Loss of function in two genes leads to synthetic lethality | Functional gene product from one gene compensates for the defective gene |
| Application | Cancer therapy targeting tumor-specific gene interactions | Gene mapping and functional analysis in genetics |
| Type of Interaction | Negative genetic interaction | Positive genetic interaction |
| Outcome | Cell death or growth inhibition | Restoration of normal growth or phenotype |
Introduction to Synthetic Lethality and Complementation
Synthetic lethality occurs when the simultaneous inactivation of two genes leads to cell death, while individual gene inactivation does not affect viability, making it a powerful tool for identifying genetic interactions and potential therapeutic targets in cancer. Complementation involves introducing a functional gene to restore the phenotype lost by a mutation, enabling researchers to confirm gene function and investigate genetic pathways. Understanding these mechanisms allows for precise manipulation of genetic networks, crucial in genetic research and drug development.
Historical Context and Discovery
Synthetic lethality was first described in the early 20th century by geneticists working with fruit flies, highlighting how the combination of two non-lethal mutations results in cell death, which provided insights into genetic interactions and functional redundancy. Complementation emerged from bacterial genetics research in the 1940s, demonstrating the ability of two mutant strains with defects in different genes to restore a wild-type phenotype when combined, a concept foundational for gene mapping and functional analysis. These discoveries shaped molecular genetics by revealing critical mechanisms of gene interaction and cellular viability.
Defining Synthetic Lethality
Synthetic lethality occurs when the simultaneous impairment of two genes leads to cell death, while the individual disruption of either gene alone is non-lethal. This concept is crucial for identifying genetic interactions that can be targeted in cancer therapy, exploiting vulnerabilities unique to malignant cells. Complementation involves the restoration of function by introducing a functional gene copy, distinguishing it from synthetic lethality's focus on combined gene defects causing lethality.
Understanding Genetic Complementation
Genetic complementation occurs when two different mutations in the heterozygous state restore the wild-type phenotype by compensating for each other's functional defects. This phenomenon indicates that the mutations affect different genes or gene functions, allowing normal cellular processes to resume. Understanding genetic complementation is crucial for mapping gene interactions and elucidating pathways involved in complex traits.
Mechanistic Differences Between the Two Concepts
Synthetic lethality occurs when the simultaneous impairment of two genes leads to cell death, while each gene's individual loss is non-lethal, highlighting a functional interaction causing essential pathway disruption. Complementation involves the restoration of function in a defective gene by introducing a wild-type allele or a compensatory gene, revealing the independent roles of genes in providing a functional product. Mechanistically, synthetic lethality uncovers dependency between parallel or redundant pathways, whereas complementation demonstrates direct gene product substitution or functional compensation at the molecular or cellular level.
Experimental Approaches for Identification
Experimental approaches for identifying synthetic lethality often involve high-throughput genetic screens using CRISPR-Cas9 or RNA interference libraries to systematically knock out or down gene pairs in cancer cell lines, revealing lethal interactions that can be therapeutically targeted. Complementation studies utilize cloning and expression of wild-type genes in mutant strains or cells to restore function, commonly assessed through phenotypic rescue assays or reporter gene activity to confirm gene function replacement. Combining genome-wide interaction maps with transcriptomic and proteomic analyses enhances precision in distinguishing synthetic lethal pairs from complementation effects in complex genetic networks.
Applications in Disease Research and Therapy
Synthetic lethality enables targeted cancer therapies by exploiting genetic interactions where the simultaneous loss of two genes results in cell death, sparing healthy cells with only one gene affected. Complementation assays are instrumental in identifying functional gene relationships and rescuing defective phenotypes, facilitating the study of genetic disorders and drug response mechanisms. Both approaches accelerate precision medicine advancements by uncovering novel therapeutic targets and biomarkers for diseases like cancer and inherited genetic conditions.
Case Studies and Notable Examples
Synthetic lethality is prominently illustrated by BRCA1/BRCA2 mutations combined with PARP inhibition in cancer therapy, where tumors deficient in DNA repair are selectively targeted. Complementation is exemplified by mitochondrial diseases where nuclear gene therapy restores function by providing wild-type gene copies to compensate for defective mitochondrial genes. Case studies of yeast genetics reveal synthetic lethality interactions helping to map functional gene networks, while complementation assays remain fundamental in identifying gene functions across various organisms.
Limitations and Challenges
Synthetic lethality faces limitations such as genetic background dependency and difficulty in identifying specific lethal gene pairs, complicating its therapeutic application. Complementation challenges include incomplete restoration of function and variability in gene expression levels that affect phenotype rescue accuracy. Both approaches require advanced genomic tools and precise functional assays to overcome off-target effects and improve prediction reliability.
Future Perspectives in Genetic Interaction Studies
Synthetic lethality holds promise for targeted cancer therapies by exploiting specific genetic vulnerabilities in tumor cells, enabling precision medicine advancements. Complementation analysis continues to be invaluable for functional genomics, aiding in the identification of gene functions and interactions with high specificity. Future genetic interaction studies are expected to integrate high-throughput sequencing and CRISPR-based screens to map complex genetic networks and accelerate the discovery of novel therapeutic targets.
Synthetic lethality Infographic
 libterm.com
libterm.com