Orthologs are genes in different species that evolved from a common ancestral gene through speciation and typically retain the same function across organisms. Understanding orthologs is crucial for comparative genomics, functional annotation, and evolutionary studies. Explore the rest of this article to learn how identifying orthologs can enhance Your insights into gene function and evolutionary relationships.
Table of Comparison
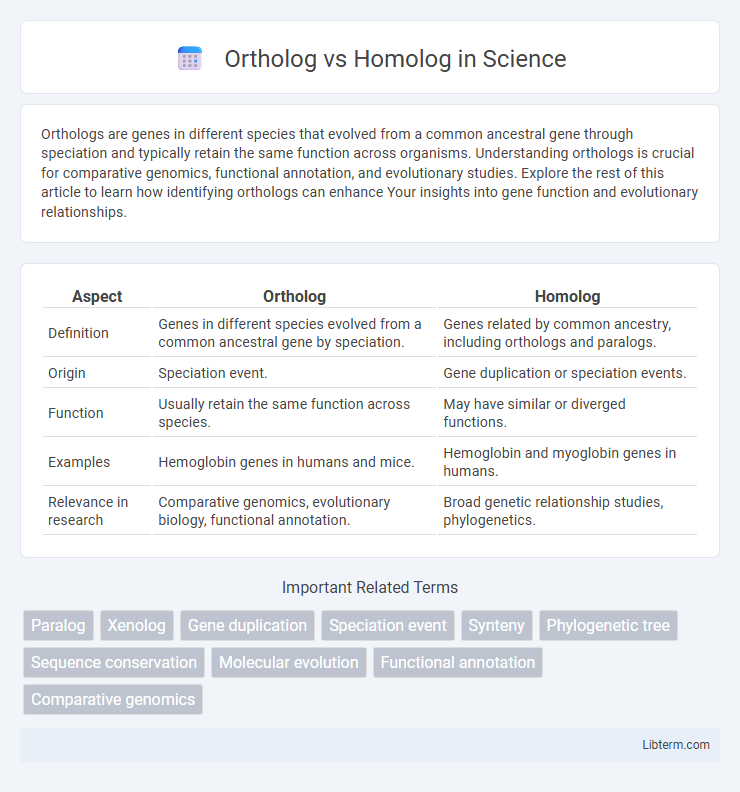
| Aspect | Ortholog | Homolog |
|---|---|---|
| Definition | Genes in different species evolved from a common ancestral gene by speciation. | Genes related by common ancestry, including orthologs and paralogs. |
| Origin | Speciation event. | Gene duplication or speciation events. |
| Function | Usually retain the same function across species. | May have similar or diverged functions. |
| Examples | Hemoglobin genes in humans and mice. | Hemoglobin and myoglobin genes in humans. |
| Relevance in research | Comparative genomics, evolutionary biology, functional annotation. | Broad genetic relationship studies, phylogenetics. |
Introduction to Orthologs and Homologs
Orthologs are genes in different species that evolved from a common ancestral gene through speciation events, retaining similar functions across species. Homologs encompass all genes related by descent from a common ancestor, including both orthologs and paralogs--genes related through duplication within a genome. Distinguishing orthologs from homologs is crucial in comparative genomics to accurately infer gene function and evolutionary relationships.
Defining Homology in Genetics
Homology in genetics refers to the existence of shared ancestry between genes or proteins in different species, indicating evolutionary relationships. Orthologs are a specific type of homologs that diverged after a speciation event, retaining the same function across species. Understanding homology helps trace gene evolution, functional conservation, and the genetic basis of traits through comparative genomics.
Understanding Orthologs: Meaning and Examples
Orthologs are genes in different species that evolved from a common ancestral gene through speciation and typically retain the same function, such as the hemoglobin genes in humans and mice. Understanding orthologs is crucial for comparative genomics and evolutionary biology because they provide insights into gene function conservation across species. Identifying orthologous genes involves sequence similarity analysis and phylogenetic methods to distinguish them from paralogs, which arise through gene duplication within the same species.
Homologs: Types and Classification
Homologs are genes related by descent from a common ancestral DNA sequence and can be classified into orthologs and paralogs based on their evolutionary history. Orthologs arise from speciation events and typically retain the same function across different species, whereas paralogs result from gene duplication events within the same genome and may evolve new functions. Understanding the distinction between these types of homologs is crucial for interpreting evolutionary relationships and functional genomics analyses.
Orthologs vs Paralogs: Key Differences
Orthologs are genes in different species that originated from a single ancestral gene through speciation, maintaining similar functions across species, while paralogs arise from gene duplication events within the same species and may evolve new functions. Understanding the distinction between orthologs and paralogs is crucial in comparative genomics and evolutionary biology for accurate functional annotation and phylogenetic analysis. Orthologs typically exhibit higher sequence conservation than paralogs, reflecting their evolutionary pressure to preserve core biological roles.
Functional Implications of Orthologs and Homologs
Orthologs are genes in different species that evolved from a common ancestral gene by speciation, typically retaining the same function across species, which makes them crucial for predicting gene function in comparative genomics. Homologs encompass both orthologs and paralogs, where paralogs arise from gene duplication events within the same species and can evolve new functions, contributing to functional diversity. Understanding the functional implications of orthologs versus homologs aids in distinguishing conserved biological roles from species-specific adaptations in evolutionary biology and molecular genetics.
Methods for Identifying Orthologs and Homologs
Methods for identifying orthologs commonly rely on sequence similarity searches using BLAST followed by reciprocal best hits analysis to pinpoint genes in different species that descended from a single ancestral gene. Homologs, encompassing orthologs and paralogs, are typically identified through phylogenetic tree reconstruction, which reveals evolutionary relationships based on sequence divergence patterns. Advanced approaches combine synteny analysis and functional annotation to improve accuracy in distinguishing orthologs from paralogs within homologous gene families.
Orthologs and Homologs in Evolutionary Biology
Orthologs are genes in different species that evolved from a common ancestral gene through speciation, retaining similar functions across organisms. Homologs encompass both orthologs and paralogs, representing genes related by descent from a common ancestor, but orthologs specifically highlight evolutionary relationships crucial for understanding functional conservation. In evolutionary biology, analyzing orthologs enables accurate inference of gene function and evolutionary history, distinguishing them from homologs derived from gene duplication events.
Applications in Comparative Genomics
Orthologs serve as crucial markers in comparative genomics for identifying genes in different species that originated from a common ancestor, enabling accurate inference of gene function across organisms. Homologs, including both orthologs and paralogs, provide a broader context for studying gene evolution, functional divergence, and phylogenetic relationships. Leveraging ortholog detection improves genome annotation, evolutionary analysis, and the understanding of conserved biological pathways across species.
Challenges and Future Directions in Ortholog and Homolog Research
Challenges in ortholog and homolog research include accurately distinguishing orthologs from paralogs due to gene duplication events and evolutionary divergence, which complicates functional predictions in comparative genomics. Advances in high-throughput sequencing and machine learning algorithms offer promising directions for improving ortholog identification accuracy and integrating phylogenomic data. Future research aims to develop standardized benchmarks and comprehensive databases to enhance the reliability of ortholog and homolog annotations across diverse species.
Ortholog Infographic
 libterm.com
libterm.com