Analog technology processes continuous signals, offering a more natural and accurate representation of real-world sounds and images compared to digital formats. It is widely used in audio equipment, film cameras, and vintage electronics for its warmth and authenticity. Explore the rest of the article to understand how analog systems impact your daily life and why they remain relevant today.
Table of Comparison
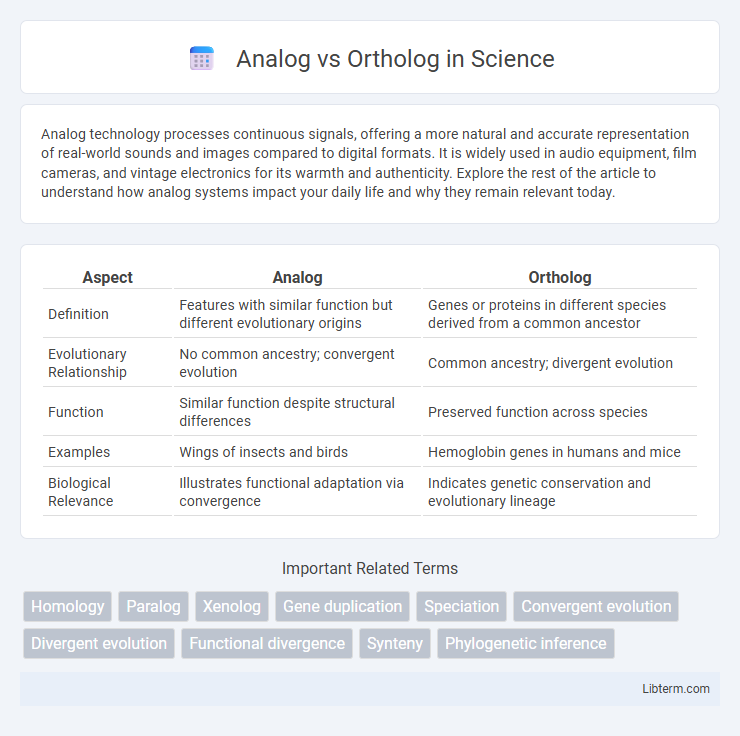
| Aspect | Analog | Ortholog |
|---|---|---|
| Definition | Features with similar function but different evolutionary origins | Genes or proteins in different species derived from a common ancestor |
| Evolutionary Relationship | No common ancestry; convergent evolution | Common ancestry; divergent evolution |
| Function | Similar function despite structural differences | Preserved function across species |
| Examples | Wings of insects and birds | Hemoglobin genes in humans and mice |
| Biological Relevance | Illustrates functional adaptation via convergence | Indicates genetic conservation and evolutionary lineage |
Introduction to Analog and Ortholog Concepts
Analogs are genes or proteins that perform similar functions across different species but do not share a common evolutionary origin, reflecting convergent evolution. Orthologs are genes in different species that evolved from a common ancestral gene through speciation and typically retain the same function. Understanding the distinction between analogs and orthologs is crucial for accurate evolutionary analysis and functional genomics studies.
Defining Analogs in Biology
Analogs in biology are structures or genes that perform similar functions in different species but have evolved independently without a common ancestor. These features arise through convergent evolution, adapting to similar environmental pressures despite distinct evolutionary pathways. Unlike orthologs, which share a common ancestral gene, analogs demonstrate functional similarity without genetic homology.
Understanding Orthologs and Their Significance
Orthologs are genes in different species that evolved from a common ancestral gene through speciation, maintaining similar functions across organisms. Understanding orthologs is essential for comparative genomics and evolutionary biology as they provide insights into gene function conservation and species divergence. Identifying orthologous genes helps in predicting gene function in newly sequenced genomes and in studying evolutionary relationships among species.
Key Differences Between Analogs and Orthologs
Analogs and orthologs differ primarily in their evolutionary origins and functions; orthologs are genes in different species that evolved from a common ancestral gene through speciation and typically retain the same function, whereas analogs arise independently in different species due to convergent evolution and often perform similar functions despite not sharing a common ancestry. Orthologous genes are crucial for studying evolutionary relationships and functional conservation across species, while analogs illustrate how similar environmental pressures can lead to similar adaptations in unrelated lineages. Understanding these distinctions aids in accurate gene annotation and evolutionary biology research.
Evolutionary Pathways of Analogs and Orthologs
Analogs arise through convergent evolution, where unrelated species independently evolve similar traits to adapt to comparable environmental pressures. Orthologs originate from a common ancestral gene and diverge following speciation events, preserving similar functions across different species. The evolutionary pathway of analogs highlights functional similarity without shared ancestry, while orthologs demonstrate genetic conservation through evolutionary divergence.
Functional Implications in Genetics
Analogs and orthologs differ significantly in their functional implications within genetics, with orthologs typically retaining similar functions across species due to their common ancestral gene origin. Analogs arise from convergent evolution and may perform similar roles but do not share a genetic lineage, leading to functional differences despite phenotypic similarities. Understanding these distinctions is crucial for interpreting gene function, evolutionary relationships, and functional genomics across diverse organisms.
Methods for Identifying Analogs and Orthologs
Methods for identifying orthologs primarily rely on sequence similarity searches using tools like BLAST along with phylogenetic tree reconstruction to confirm evolutionary relationships. Analog identification depends more on structural and functional analyses, comparing protein folds and biological roles rather than direct sequence homology. Combining computational approaches such as sequence alignment, domain architecture comparison, and functional assays enhances the accurate distinction between orthologous and analogous genes.
Relevance to Comparative Genomics
Orthologs are genes in different species that evolved from a common ancestral gene by speciation, retaining similar functions, making them crucial for inferring evolutionary relationships and functional conservation in comparative genomics. Analogs are genes or proteins that perform similar functions but evolved independently, providing insights into convergent evolution rather than direct lineage comparison. Distinguishing orthologs from analogs enhances the accuracy of phylogenetic analysis and functional annotation across diverse genomes.
Applications in Molecular Biology Research
Orthologs serve as critical tools in molecular biology for inferring gene function and evolutionary relationships across different species due to their conserved sequences and roles. Analogs, though not derived from common ancestry, provide insights into convergent evolution and functional adaptation by exhibiting similar biological functions despite structural differences. These distinctions enable precise gene annotation, functional prediction, and evolutionary studies, enhancing comparative genomics and protein engineering applications.
Future Perspectives and Challenges
Future perspectives in studying analogs and orthologs emphasize advanced computational tools and machine learning to enhance functional annotation accuracy across diverse genomes. Challenges include resolving evolutionary divergence complexities and distinguishing functional convergence in analogs versus conserved mechanisms in orthologs. Integrating multi-omics data and improving sequence homology detection will be pivotal for precise gene function prediction and evolutionary biology research.
Analog Infographic
 libterm.com
libterm.com