Phylogenetics explores the evolutionary relationships among species through genetic, morphological, and molecular data analysis, uncovering the shared ancestry and divergence of life forms. This field employs phylogenetic trees to visualize these connections and trace lineage patterns over time. Discover how understanding phylogenetics can enhance your grasp of biodiversity by reading the rest of the article.
Table of Comparison
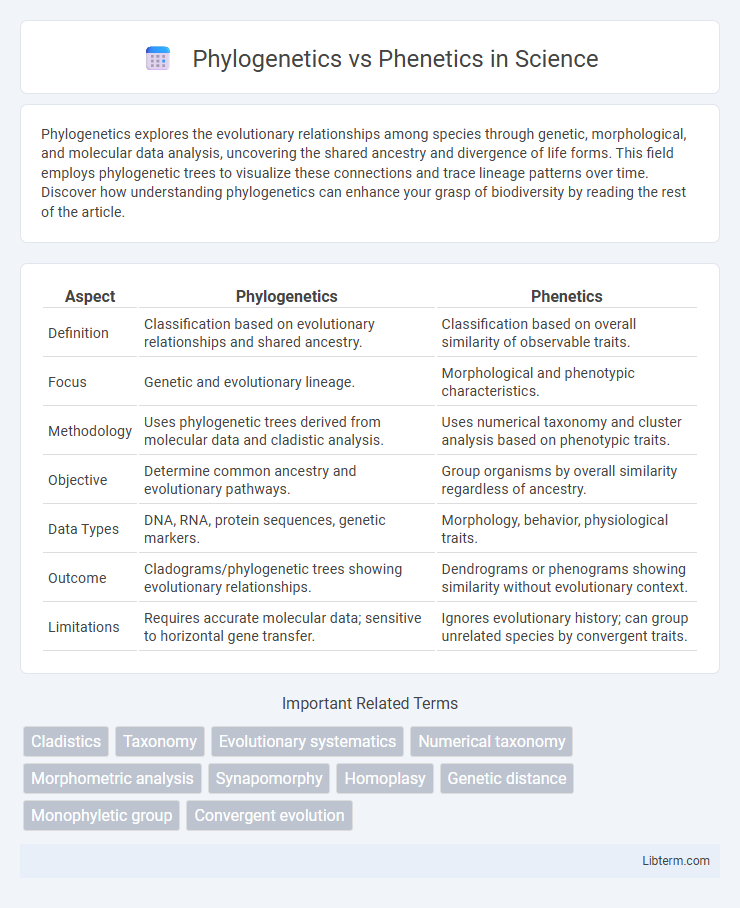
| Aspect | Phylogenetics | Phenetics |
|---|---|---|
| Definition | Classification based on evolutionary relationships and shared ancestry. | Classification based on overall similarity of observable traits. |
| Focus | Genetic and evolutionary lineage. | Morphological and phenotypic characteristics. |
| Methodology | Uses phylogenetic trees derived from molecular data and cladistic analysis. | Uses numerical taxonomy and cluster analysis based on phenotypic traits. |
| Objective | Determine common ancestry and evolutionary pathways. | Group organisms by overall similarity regardless of ancestry. |
| Data Types | DNA, RNA, protein sequences, genetic markers. | Morphology, behavior, physiological traits. |
| Outcome | Cladograms/phylogenetic trees showing evolutionary relationships. | Dendrograms or phenograms showing similarity without evolutionary context. |
| Limitations | Requires accurate molecular data; sensitive to horizontal gene transfer. | Ignores evolutionary history; can group unrelated species by convergent traits. |
Introduction to Phylogenetics and Phenetics
Phylogenetics studies evolutionary relationships among species by analyzing genetic, morphological, and molecular data to construct phylogenetic trees that represent ancestral lineages. Phenetics, also known as numerical taxonomy, classifies organisms based on overall similarity, using quantitative measures without necessarily considering evolutionary history. Both approaches utilize statistical methods, but phylogenetics emphasizes shared ancestry while phenetics focuses on phenotypic resemblance.
Defining Phylogenetics
Phylogenetics studies the evolutionary relationships among species by analyzing genetic, morphological, or molecular data to construct a phylogenetic tree. It emphasizes common ancestry and descent patterns, using methods like cladistics to infer shared derived characteristics (synapomorphies). This approach contrasts with phenetics, which groups organisms based on overall similarity without considering evolutionary history.
Defining Phenetics
Phenetics, also known as numerical taxonomy, classifies organisms based on overall similarity in observable traits without considering evolutionary relationships. It uses quantitative methods and computer algorithms to analyze multiple characteristics, creating phenograms that cluster taxa by resemblance. This approach contrasts with phylogenetics, which reconstructs evolutionary history through shared derived characters and genetic data.
Historical Development of Both Approaches
Phylogenetics emerged in the mid-20th century with roots in evolutionary theory, emphasizing the reconstruction of evolutionary relationships based on shared ancestry and cladistic methods pioneered by Willi Hennig. Phenetics developed earlier in the 1950s and 1960s, focusing on overall similarity among organisms using numerical taxonomy and statistical techniques without considering evolutionary history. The historical development of phylogenetics marked a shift towards evolutionary context, while phenetics prioritized quantitative data to classify species based solely on observable traits.
Methodologies in Phylogenetic Analysis
Phylogenetic analysis primarily utilizes cladistics, which focuses on shared derived characters (synapomorphies) to reconstruct evolutionary relationships and generate a branching tree known as a cladogram. Methods include maximum parsimony, maximum likelihood, and Bayesian inference, each employing statistical models to estimate the most probable phylogenetic tree from molecular or morphological data. Phenetics, in contrast, relies on overall similarity measured by distance-based methods without distinguishing between ancestral and derived traits, often resulting in phenograms that do not reflect evolutionary history.
Techniques Used in Phenetic Classification
Phenetic classification relies on numerical taxonomy techniques that quantify overall similarity between organisms based on observable traits, often using algorithms such as cluster analysis and principal component analysis. Morphological, biochemical, and molecular data serve as the primary inputs for these techniques, enabling the grouping of species by phenotypic attributes without consideration of evolutionary relationships. This approach contrasts with phylogenetics, which prioritizes ancestral lineage reconstruction through genetic sequence alignment and cladistic methods.
Key Differences Between Phylogenetics and Phenetics
Phylogenetics classifies organisms based on evolutionary relationships and common ancestry using genetic, molecular, and morphological data, while phenetics groups organisms solely on overall similarity without considering evolutionary history. Phylogenetics employs cladistic methods and produces cladograms reflecting evolutionary pathways, whereas phenetics relies on phenotypic traits and numerical taxonomy techniques to cluster organisms. The key difference lies in phylogenetics emphasizing ancestral lineage and evolutionary divergence, whereas phenetics focuses on observable characteristics and overall resemblance among species.
Advantages and Limitations of Each Approach
Phylogenetics offers the advantage of reconstructing evolutionary relationships based on genetic data, providing insights into ancestral lineages and speciation events, but its limitation lies in the potential for homoplasy and incomplete lineage sorting to obscure true relationships. Phenetics, which classifies organisms solely by overall similarity using measurable traits, benefits from simplicity and the inclusion of extensive morphological data, yet it cannot distinguish convergent traits from shared ancestry, often leading to misleading groupings. Both approaches rely on different data types and methods, making the integration of phylogenetic and phenetic data essential for a comprehensive understanding of organismal diversity and evolutionary history.
Applications in Modern Taxonomy and Systematics
Phylogenetics leverages evolutionary relationships through genetic data to construct cladograms that accurately reflect species divergence, making it essential for modern taxonomy and systematics in discerning ancestral lineages. Phenetics, based on overall morphological similarities, is useful for initial species grouping and quick identification but may overlook evolutionary history, limiting its application in resolving deep phylogenetic relationships. Integrating phylogenetic methodologies enhances classification systems by providing a robust framework for understanding biodiversity and speciation events in contemporary biological research.
Future Trends in Classification Science
Future trends in classification science emphasize integrating phylogenetics with phenetics to enhance evolutionary understanding and trait analysis using advanced machine learning algorithms and big data analytics. High-throughput sequencing technologies combined with computational modeling are driving more precise and dynamic classification systems based on genetic, morphological, and ecological data. The emergence of integrative taxonomy aims to unify phenetic similarity measures with phylogenetic inference, fostering more robust and predictive models of biodiversity evolution.
Phylogenetics Infographic
 libterm.com
libterm.com