Junk DNA, once thought to be useless genetic material, actually plays critical roles in regulating gene expression and maintaining genome stability. Understanding the functions of these non-coding regions helps reveal how complex biological processes are controlled at the molecular level. Discover how this hidden portion of your genome influences health and development by reading the rest of the article.
Table of Comparison
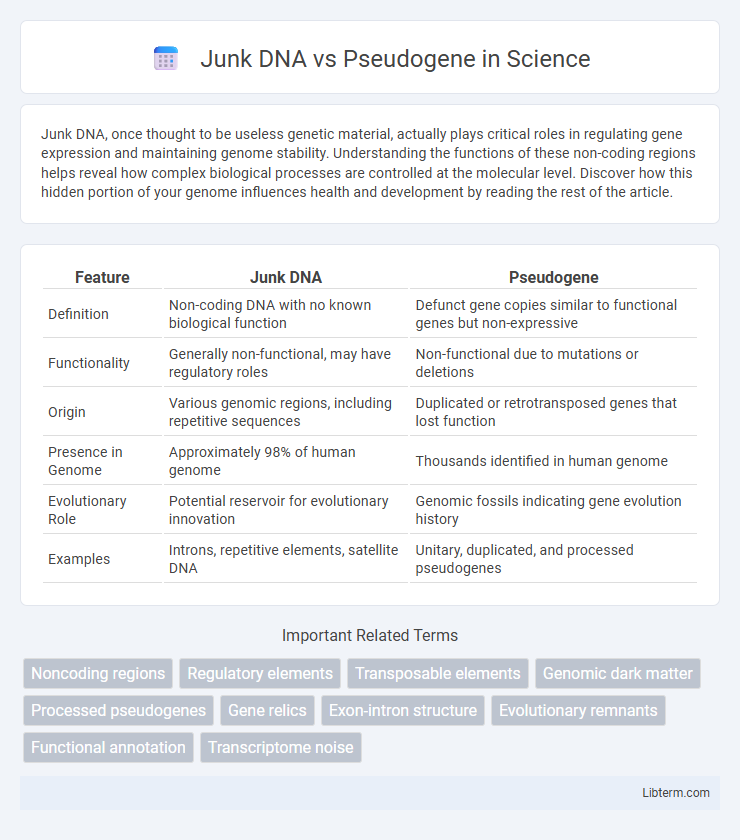
| Feature | Junk DNA | Pseudogene |
|---|---|---|
| Definition | Non-coding DNA with no known biological function | Defunct gene copies similar to functional genes but non-expressive |
| Functionality | Generally non-functional, may have regulatory roles | Non-functional due to mutations or deletions |
| Origin | Various genomic regions, including repetitive sequences | Duplicated or retrotransposed genes that lost function |
| Presence in Genome | Approximately 98% of human genome | Thousands identified in human genome |
| Evolutionary Role | Potential reservoir for evolutionary innovation | Genomic fossils indicating gene evolution history |
| Examples | Introns, repetitive elements, satellite DNA | Unitary, duplicated, and processed pseudogenes |
Introduction to Noncoding DNA
Noncoding DNA constitutes over 98% of the human genome and includes sequences such as junk DNA and pseudogenes, which do not code for proteins but play varied roles in genome regulation. Junk DNA refers to regions initially thought to lack function, though many contain regulatory elements essential for gene expression and chromosomal architecture. Pseudogenes are former genes rendered nonfunctional through mutations but may influence gene regulation by acting as decoys for microRNAs or producing noncoding RNAs.
Defining Junk DNA
Junk DNA refers to non-coding regions of the genome once considered functionally inert, comprising approximately 98-99% of human DNA. These sequences include repetitive elements, introns, and regulatory regions that do not code for proteins but may have regulatory or structural roles. Unlike pseudogenes, which are defective gene copies with recognizable homology to functional genes, junk DNA encompasses a broader array of non-functional or poorly understood genomic segments.
Understanding Pseudogenes
Pseudogenes are non-functional segments of DNA that resemble functional genes but have lost their protein-coding ability due to mutations or deletions, distinguishing them from junk DNA, which lacks any identifiable gene-like characteristics. Unlike junk DNA, pseudogenes often arise from gene duplication or retrotransposition events and can provide evolutionary insights or regulatory functions in gene expression. Understanding pseudogenes involves studying their sequences, origins, and potential roles in genomic regulation and disease mechanisms.
Origins of Junk DNA
Junk DNA primarily originates from repetitive sequences, transposable elements, and nonfunctional remnants of ancient viral insertions, making up nearly 50-60% of the human genome. Pseudogenes arise from duplicated or mutated genes that have lost their ability to code for functional proteins but retain sequence similarity to their ancestral genes. Understanding the distinct origins of junk DNA and pseudogenes sheds light on genome evolution and regulatory complexity in eukaryotic organisms.
Formation and Types of Pseudogenes
Pseudogenes form from previously functional genes that accumulate mutations, rendering them noncoding and unable to produce functional proteins, while junk DNA comprises noncoding DNA with unclear or no function. Types of pseudogenes include processed pseudogenes, which arise from reverse-transcribed mRNA inserted back into the genome without regulatory elements, and non-processed pseudogenes, formed by gene duplication followed by disabling mutations. Unitary pseudogenes result from inactivation of a single functional gene without duplication, differentiating them from other pseudogene classes based on their origin and genomic context.
Functional Roles of Junk DNA
Junk DNA, once thought to be non-functional, includes regulatory elements, non-coding RNAs, and sequences influencing chromatin structure, playing crucial roles in gene expression and genome stability. Pseudogenes, a subset of Junk DNA, are gene copies that have lost protein-coding ability but can regulate their functional counterparts through mechanisms like RNA interference. Understanding these elements highlights that Junk DNA contributes to cellular processes beyond protein coding, affecting genetic regulation and evolution.
Potential Functions of Pseudogenes
Pseudogenes, once considered nonfunctional genomic fossils, exhibit potential regulatory roles in gene expression by acting as microRNA sponges or sources of small interfering RNAs, influencing the stability and translation of messenger RNAs. Unlike junk DNA, pseudogenes can modulate cellular processes through their transcriptional activity, contributing to gene regulation networks and phenotypic diversity. Emerging research highlights pseudogenes' involvement in disease mechanisms and evolutionary adaptation, emphasizing their functional relevance beyond genomic redundancy.
Junk DNA vs Pseudogene: Key Differences
Junk DNA consists of non-coding sequences in the genome once thought to have no function, whereas pseudogenes are remnants of once-functional genes that have lost their protein-coding ability due to mutations. Unlike junk DNA, pseudogenes often share significant sequence similarity with functional genes and can arise through gene duplication or retrotransposition. The key difference lies in their origin and potential regulatory roles, with pseudogenes sometimes influencing gene expression, while junk DNA is largely considered genetically inert.
Research Advances in Noncoding Genomics
Recent research advances in noncoding genomics have redefined the understanding of junk DNA and pseudogenes, revealing their functional roles in gene regulation and evolutionary processes. High-throughput sequencing and CRISPR-based functional assays have identified regulatory elements and noncoding RNAs derived from previously labeled junk DNA, demonstrating diverse contributions to cellular mechanisms. Pseudogenes, once considered genomic fossils, are now recognized for their regulatory potential through RNA interference and competitive endogenous RNA networks, highlighting their significance in disease and developmental biology.
Implications for Genetics and Evolution
Junk DNA, once considered nonfunctional, consists primarily of repetitive sequences and non-coding regions that may regulate gene expression and genome architecture, whereas pseudogenes are degraded gene copies reflecting evolutionary remnants of ancestral genes. Research into pseudogenes reveals their potential roles in gene regulation and genome plasticity, challenging the misconception that non-coding DNA lacks functionality. Understanding the distinction between junk DNA and pseudogenes enhances insights into genetic variation, evolutionary mechanisms, and the complexity of genomic regulation.
Junk DNA Infographic
 libterm.com
libterm.com