Paralogy refers to the presence of genes within the same organism that have evolved by duplication and subsequently diverged in function. These paralogous genes play crucial roles in genetic innovation and complexity across species. Explore the rest of the article to understand how paralogy impacts genetics and evolutionary biology.
Table of Comparison
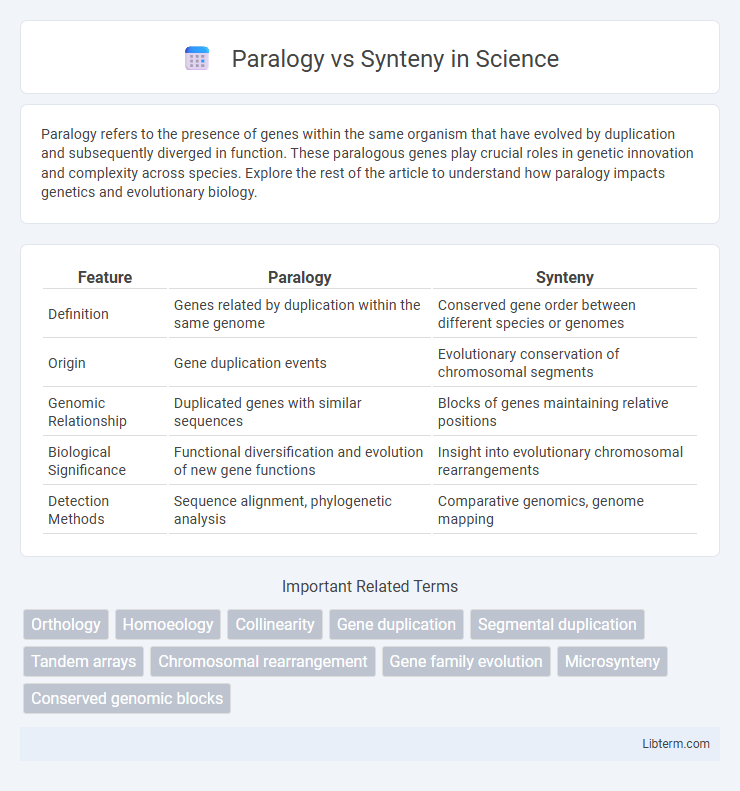
| Feature | Paralogy | Synteny |
|---|---|---|
| Definition | Genes related by duplication within the same genome | Conserved gene order between different species or genomes |
| Origin | Gene duplication events | Evolutionary conservation of chromosomal segments |
| Genomic Relationship | Duplicated genes with similar sequences | Blocks of genes maintaining relative positions |
| Biological Significance | Functional diversification and evolution of new gene functions | Insight into evolutionary chromosomal rearrangements |
| Detection Methods | Sequence alignment, phylogenetic analysis | Comparative genomics, genome mapping |
Understanding Paralogy: Definition and Significance
Paralogy refers to gene duplication within a genome resulting in gene copies that have evolved new functions, playing a crucial role in genomic innovation and complexity. These paralogous genes arise through duplication events and contribute to functional diversification by acquiring unique roles that enhance adaptability. Understanding paralogy is essential for interpreting evolutionary relationships and the functional interplay between genes within and across species.
What is Synteny? An Overview
Synteny refers to the conservation of blocks of genes on chromosomes across different species, indicating shared ancestry and evolutionary relationships. It highlights the physical co-localization of genes in chromosomal segments between species, which can be crucial for comparative genomics and evolutionary biology studies. Understanding synteny helps researchers trace genome rearrangements, gene duplications, and the evolutionary history of genomes.
Paralogy vs Synteny: Key Differences
Paralogy and synteny refer to different genomic concepts important in evolutionary biology and genomics. Paralogy describes genes related by duplication within a genome, resulting in gene families with similar sequences but potentially diverse functions. Synteny refers to the conservation of gene order on chromosomes across species, highlighting regions where gene clusters remain unchanged through evolutionary time, indicating shared ancestry and structural genome organization.
Evolutionary Implications of Paralogy
Paralogy arises from gene duplication events, leading to the presence of gene copies within the same genome that can evolve new functions and contribute to organismal complexity. Unlike synteny, which refers to conserved gene order across species, paralogous genes highlight evolutionary processes such as neofunctionalization and subfunctionalization. Understanding paralogy is crucial for tracing gene family expansions and adaptive innovation in evolutionary biology.
The Role of Synteny in Genome Organization
Synteny refers to the conserved order of genes on chromosomes across different species, playing a crucial role in maintaining genome organization and facilitating the study of evolutionary relationships. Unlike paralogy, which involves gene duplication events within a genome, synteny highlights chromosomal regions that remain structurally intact over time. The preservation of syntenic blocks aids in identifying homologous genes and understanding genome rearrangements critical to species divergence.
Methods for Detecting Paralogy
Paralogy detection methods primarily rely on sequence similarity searches using BLAST or DIAMOND, followed by clustering algorithms like OrthoMCL or Markov clustering to group homologous genes. Phylogenetic tree construction further distinguishes paralogous genes from orthologs by analyzing gene duplication events within gene families. Comparative genomics approaches integrate synteny information to validate paralogs by examining conserved gene order across related species.
Approaches to Identifying Syntenic Regions
Identifying syntenic regions involves comparative genomic approaches such as sequence alignment tools, including BLAST and LASTZ, to detect conserved gene order across different species. Computational algorithms like MCScanX utilize gene collinearity and homology data to map synteny blocks, emphasizing the preservation of gene clusters rather than mere sequence similarity. These methods contrast with paralogy identification, which focuses on gene duplication events within the same genome rather than cross-species chromosomal conservation.
Functional Consequences of Paralogous Genes
Paralogous genes arise from gene duplication events and often retain similar but distinct functions, enabling evolutionary innovation and increased organismal complexity. These genes can undergo subfunctionalization, where each paralog specializes in a subset of the original gene's role, or neofunctionalization, where one paralog acquires a novel function. The functional divergence of paralogs impacts biochemical pathways, gene regulatory networks, and phenotypic traits, contributing to adaptive evolution and species diversification.
Applications of Synteny in Comparative Genomics
Synteny analysis enables the identification of conserved gene order across different species, facilitating the study of evolutionary relationships and chromosomal rearrangements. It supports genome annotation by transferring functional information between orthologous genes and improves the understanding of genome architecture and speciation processes. Applications include identifying genomic regions involved in adaptation, tracing gene family expansions, and reconstructing ancestral genomes in comparative genomics research.
Future Perspectives: Integrating Paralogy and Synteny Analysis
Future perspectives in genomic research emphasize integrating paralogy and synteny analysis to enhance understanding of genome evolution and functional genomics. Combining paralogy, which identifies gene duplications within genomes, with synteny, the conserved order of genes across species, enables more precise annotation of duplicated genes and improves detection of evolutionary rearrangements. Advances in computational tools and high-throughput sequencing data will facilitate comprehensive integration, driving discoveries in comparative genomics and molecular evolution.
Paralogy Infographic
 libterm.com
libterm.com