Evolutionary taxonomy classifies organisms based on their shared ancestry and evolutionary relationships, emphasizing genetic lineage over superficial similarities. This approach helps clarify the tree of life by grouping species according to common descent and evolutionary traits. Explore the rest of this article to understand how evolutionary taxonomy reshapes our view of biodiversity and species classification.
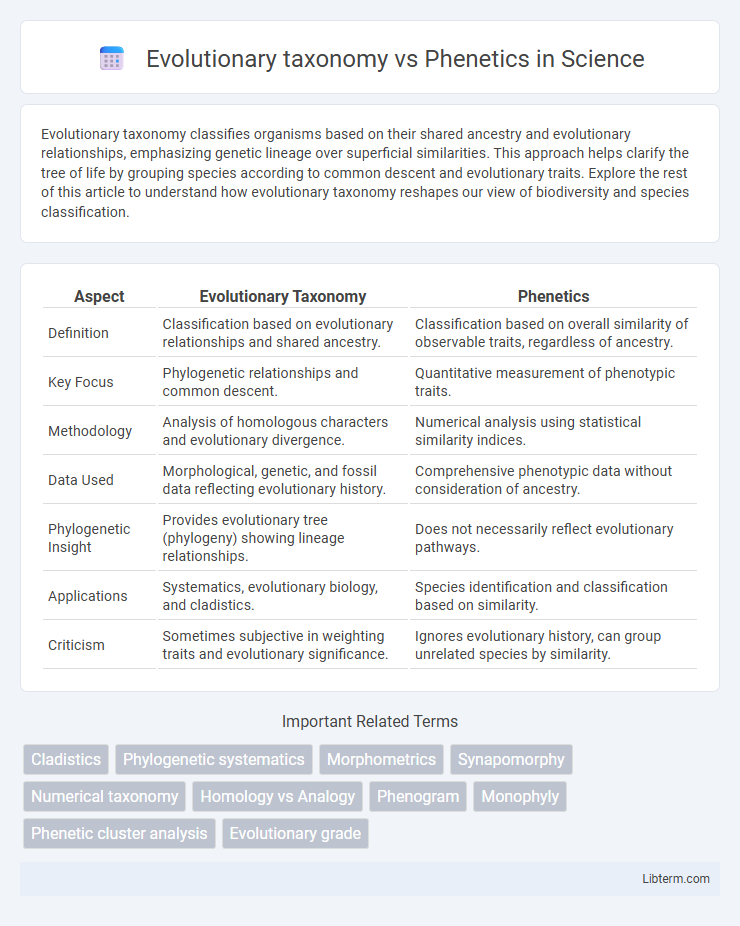
Table of Comparison
| Aspect | Evolutionary Taxonomy | Phenetics |
|---|---|---|
| Definition | Classification based on evolutionary relationships and shared ancestry. | Classification based on overall similarity of observable traits, regardless of ancestry. |
| Key Focus | Phylogenetic relationships and common descent. | Quantitative measurement of phenotypic traits. |
| Methodology | Analysis of homologous characters and evolutionary divergence. | Numerical analysis using statistical similarity indices. |
| Data Used | Morphological, genetic, and fossil data reflecting evolutionary history. | Comprehensive phenotypic data without consideration of ancestry. |
| Phylogenetic Insight | Provides evolutionary tree (phylogeny) showing lineage relationships. | Does not necessarily reflect evolutionary pathways. |
| Applications | Systematics, evolutionary biology, and cladistics. | Species identification and classification based on similarity. |
| Criticism | Sometimes subjective in weighting traits and evolutionary significance. | Ignores evolutionary history, can group unrelated species by similarity. |
Introduction to Evolutionary Taxonomy and Phenetics
Evolutionary taxonomy classifies organisms based on their phylogenetic relationships and shared evolutionary ancestry, emphasizing homology and derived traits to reflect evolutionary history. Phenetics, or numerical taxonomy, groups organisms solely by overall similarity in measurable characteristics, often using statistical methods without considering evolutionary relationships. This distinction highlights evolutionary taxonomy's goal to depict lineage connections, whereas phenetics prioritizes objective quantification of observable traits.
Historical Background and Development
Evolutionary taxonomy emerged in the early 20th century, integrating Darwinian principles of common ancestry and evolutionary relationships to classify organisms based on shared traits and phylogenetic lineage. Phenetics, developed in the mid-20th century by Robert R. Sokal and Peter H. A. Sneath, emphasized numerical methods and overall similarity rather than evolutionary history to group organisms using quantitative morphological and genetic data. The historical development of evolutionary taxonomy reflected a shift toward phylogenetic reasoning, while phenetics represented a more data-driven, algorithmic approach to classification before cladistics gained prominence.
Core Principles of Evolutionary Taxonomy
Evolutionary taxonomy classifies organisms based on shared ancestry and evolutionary relationships, integrating both genetic lineage and morphological traits. It emphasizes common descent and evolutionary divergence, prioritizing monophyletic groups while accommodating paraphyletic groups for practical classification. Phenetics, by contrast, classifies organisms solely on overall similarity without considering evolutionary history, relying primarily on measurable phenotypic traits.
Core Principles of Phenetics
Phenetics classifies organisms based on overall similarity, typically using quantitative measures of morphological or genetic traits without explicitly considering evolutionary relationships. It emphasizes the use of numerical methods and statistical analysis to cluster organisms into groups, relying on phenotypic characters as data points for classification. Core principles of phenetics include the assumption that similarity reflects relatedness and the objective, reproducible nature of its algorithms to generate classifications.
Major Differences Between Evolutionary Taxonomy and Phenetics
Evolutionary taxonomy classifies organisms based on both evolutionary relationships and morphological characteristics, emphasizing common ancestry and phylogenetic context. Phenetics relies solely on overall similarity in observable traits without considering evolutionary lineage or genetic information. The major difference lies in evolutionary taxonomy incorporating phylogenetic history, while phenetics focuses on quantifying phenotypic resemblance without evolutionary interpretation.
Methodologies and Analytical Approaches
Evolutionary taxonomy employs phylogenetic methods that analyze shared derived characteristics to reconstruct evolutionary relationships based on common ancestry, using cladistic analysis and molecular data for more accurate tree-building. Phenetics relies on overall similarity by quantifying numerous morphological and biochemical traits, applying numerical techniques such as cluster analysis and principal component analysis to group organisms without regard to evolutionary history. The distinct methodologies reflect evolutionary taxonomy's focus on inferential evolutionary processes, while phenetics emphasizes objective, quantitative measurement of phenotypic traits to categorize biodiversity.
Advantages and Limitations of Evolutionary Taxonomy
Evolutionary taxonomy integrates phylogenetic relationships and morphological characteristics to classify organisms, offering a more comprehensive understanding of evolutionary history and ancestral traits. Its advantages include reflecting both genetic lineage and phenotypic divergence, which aids in tracing evolutionary processes and adaptation patterns. Limitations involve potential subjectivity in weighing traits and the challenge of incorporating horizontal gene transfer, which can obscure clear lineage distinctions compared to strictly phenetic approaches based on measurable similarities.
Advantages and Limitations of Phenetics
Phenetics classifies organisms based on overall similarity, primarily using quantitative measurements and numerical methods, which allows for objective and reproducible classification. Its advantage lies in the ability to analyze large datasets and produce clear visualizations like cluster diagrams, aiding in identifying phenotypic patterns. However, phenetics ignores evolutionary relationships and homology, potentially grouping organisms with convergent traits rather than true phylogenetic kinship, limiting its utility for understanding evolutionary history.
Real-world Applications and Case Studies
Evolutionary taxonomy, which classifies organisms based on shared ancestry and phylogenetic relationships, is extensively applied in conservation biology to identify evolutionary significant units for species preservation. Phenetics, relying on overall morphological similarity without considering evolutionary history, proves useful in paleontology where fossil records lack genetic information, aiding in the classification of extinct species. Case studies in microbial taxonomy demonstrate how evolutionary taxonomy informs the development of targeted antibiotics through phylogenetic analysis, while phenetic approaches streamline identification processes in ecological surveys by clustering organisms based on observable traits.
Future Perspectives in Biological Classification
Future perspectives in biological classification increasingly emphasize integration of evolutionary taxonomy and phenetics to enhance accuracy in species identification and phylogenetic relationships. Advances in genomic sequencing and computational tools enable deeper analysis of phenotypic traits alongside genetic data, fostering more robust, multidimensional classification systems. This synthesis promises to resolve taxonomic ambiguities by combining phenetic similarity with evolutionary lineage information for a comprehensive understanding of biodiversity.
Evolutionary taxonomy Infographic
 libterm.com
libterm.com