DNA transposons are mobile genetic elements that can change their position within the genome through a "cut and paste" mechanism, impacting genome stability and evolution. These sequences encode transposase enzymes that facilitate their excision and integration into new genomic locations, often influencing gene function and regulation. Discover how DNA transposons affect genetic diversity and what implications they hold for your understanding of molecular biology in the rest of this article.
Table of Comparison
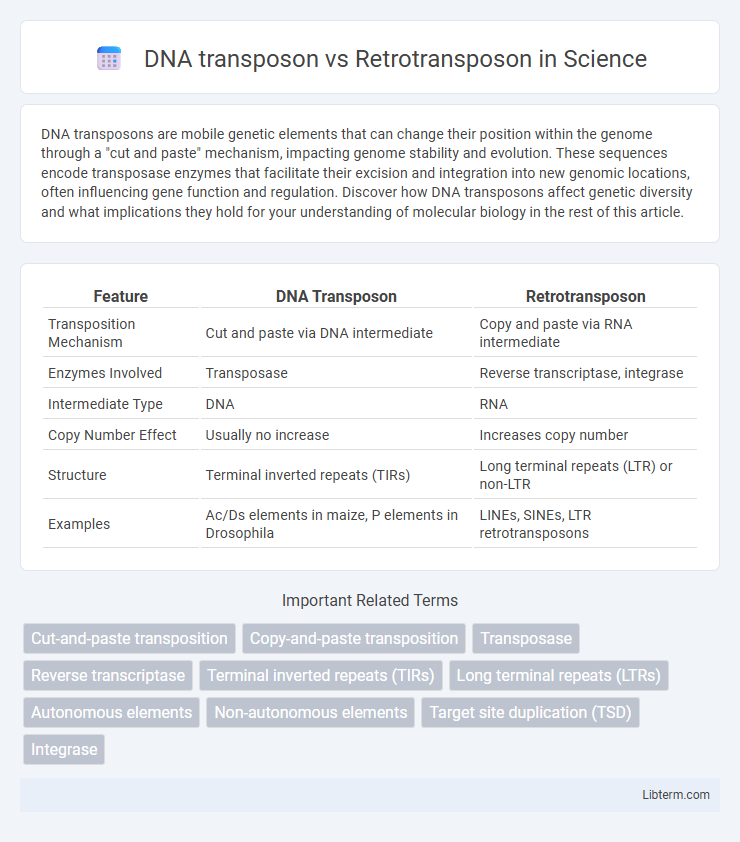
| Feature | DNA Transposon | Retrotransposon |
|---|---|---|
| Transposition Mechanism | Cut and paste via DNA intermediate | Copy and paste via RNA intermediate |
| Enzymes Involved | Transposase | Reverse transcriptase, integrase |
| Intermediate Type | DNA | RNA |
| Copy Number Effect | Usually no increase | Increases copy number |
| Structure | Terminal inverted repeats (TIRs) | Long terminal repeats (LTR) or non-LTR |
| Examples | Ac/Ds elements in maize, P elements in Drosophila | LINEs, SINEs, LTR retrotransposons |
Introduction to Transposable Elements
Transposable elements are genetic sequences that can move within a genome, categorized mainly into DNA transposons and retrotransposons. DNA transposons relocate through a "cut-and-paste" mechanism involving a transposase enzyme, whereas retrotransposons move via an RNA intermediate through a "copy-and-paste" process using reverse transcriptase. These mobile genetic elements contribute to genome evolution, variation, and can influence gene expression and genomic stability.
Overview of DNA Transposons
DNA transposons, also known as "jumping genes," move within the genome via a cut-and-paste mechanism facilitated by the transposase enzyme, allowing direct excision and reinsertion. These elements differ from retrotransposons, which transpose through an RNA intermediate using reverse transcriptase. DNA transposons contribute to genetic diversity and genome evolution by creating mutations, gene duplications, and chromosomal rearrangements.
Mechanism of DNA Transposon Movement
DNA transposons move via a "cut-and-paste" mechanism where the transposase enzyme excises the element from one genomic location and integrates it into another, creating target site duplications at the insertion point. This movement does not involve an RNA intermediate, distinguishing it from retrotransposons, which replicate through an RNA intermediate using reverse transcriptase. DNA transposon mobility impacts genome structure by causing insertions, deletions, and chromosomal rearrangements during transposition events.
Overview of Retrotransposons
Retrotransposons are genetic elements that replicate through an RNA intermediate, using a "copy and paste" mechanism involving reverse transcription to insert copies of themselves throughout the genome. They are classified mainly into long terminal repeat (LTR) retrotransposons, which resemble retroviruses, and non-LTR retrotransposons, such as LINEs and SINEs, which lack LTRs but contribute significantly to genomic variation and evolution. These elements can impact genome structure and function by creating mutations, altering gene expression, and facilitating genomic rearrangements.
Mechanism of Retrotransposon Migration
Retrotransposons migrate through a "copy-and-paste" mechanism involving reverse transcription, where an RNA intermediate is synthesized from the retrotransposon DNA, followed by reverse transcription into cDNA, which integrates into a new genomic location. This process relies on the enzyme reverse transcriptase, encoded by the retrotransposon itself, facilitating the synthesis of DNA from RNA. Unlike DNA transposons that use a "cut-and-paste" mechanism without RNA intermediates, retrotransposons increase their copy number within the genome during transposition.
Structural Differences Between DNA Transposons and Retrotransposons
DNA transposons consist of inverted repeat sequences flanking a transposase gene, enabling a cut-and-paste mechanism for mobility. Retrotransposons feature long terminal repeats (LTRs) or non-LTR elements with reverse transcriptase and integrase genes, facilitating an RNA intermediate that is reverse-transcribed into DNA for integration. These structural differences underpin DNA transposons' direct excision and reinsertion contrasting with retrotransposons' copy-and-paste replication via an RNA intermediate.
Genomic Impact and Evolutionary Significance
DNA transposons, which move via a "cut-and-paste" mechanism, contribute to genome plasticity by creating insertions and deletions that can disrupt gene function or regulatory regions, influencing genome architecture and evolution. Retrotransposons propagate through an RNA intermediate using a "copy-and-paste" mechanism, leading to genome expansion and increased genetic diversity by generating multiple copies that can induce mutations and chromosomal rearrangements. Both transposon types drive evolutionary innovation by facilitating gene duplication, exon shuffling, and regulatory network modifications, significantly shaping genome structure and adaptability across species.
Regulation and Control of Transposon Activity
DNA transposons are regulated primarily through host-encoded proteins that inhibit transposase expression or activity, ensuring low transposition rates to maintain genomic stability. Retrotransposons rely on epigenetic mechanisms such as DNA methylation and histone modification to suppress their transcription and prevent excessive mobilization. Both transposon types are subject to RNA interference pathways that degrade transposon-derived RNAs, limiting their potential for harmful insertions.
Biological Consequences and Disease Associations
DNA transposons move through a "cut-and-paste" mechanism, causing genomic insertions that can disrupt gene function and lead to mutations linked to cancers and genetic disorders. Retrotransposons replicate via an RNA intermediate, increasing their copy number and contributing to genome instability, with strong associations to neurodegenerative diseases and autoimmune conditions. Both types of transposable elements influence genome evolution, but their mobilization can promote harmful chromosomal rearrangements and aberrant gene expression patterns.
Comparative Summary: DNA Transposon vs Retrotransposon
DNA transposons move via a "cut-and-paste" mechanism using a transposase enzyme to excise and reintegrate DNA segments, whereas retrotransposons utilize a "copy-and-paste" approach through RNA intermediates and reverse transcriptase to insert copies into new genomic locations. DNA transposons tend to have terminal inverted repeats and generate target site duplications upon insertion, while retrotransposons often feature long terminal repeats (LTRs) or lack LTRs in non-LTR types like LINEs and SINEs. The replication mode of retrotransposons contributes to genome expansion more significantly than DNA transposons, which typically do not increase copy number during transposition.
DNA transposon Infographic
 libterm.com
libterm.com