Satellite DNA consists of repetitive, non-coding sequences found primarily in centromeric and heterochromatic regions of chromosomes, playing a crucial role in chromosome structure and stability. These highly repetitive sequences vary among species and contribute to genomic organization and function, often influencing genetic regulation and evolution. Explore the rest of the article to understand how satellite DNA impacts your genetic makeup and its significance in molecular biology.
Table of Comparison
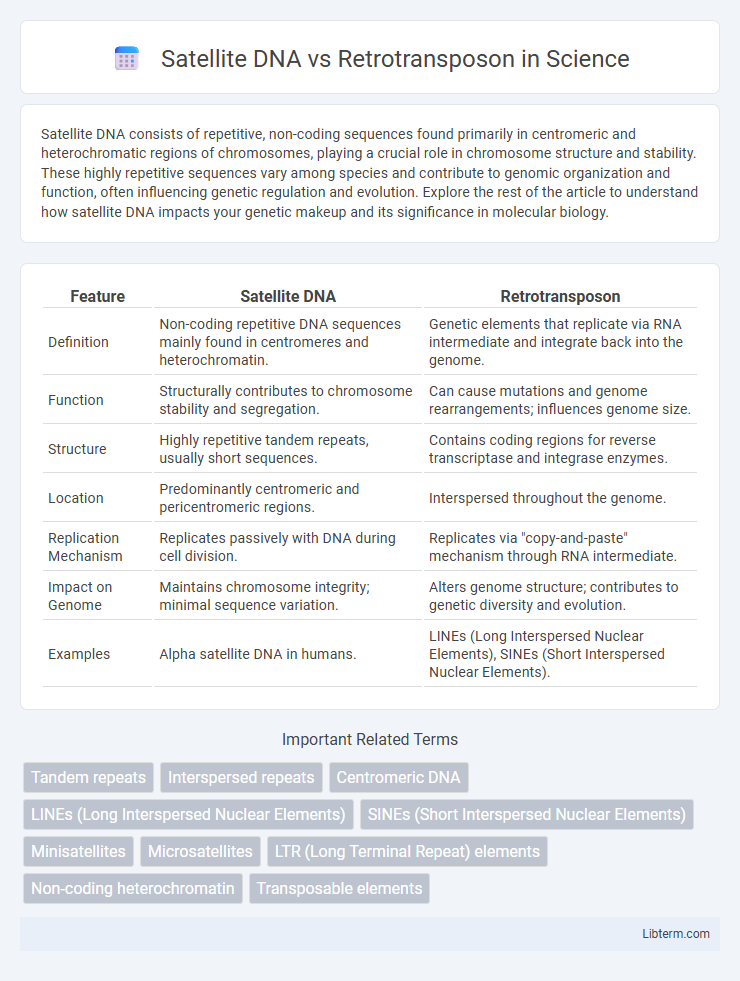
| Feature | Satellite DNA | Retrotransposon |
|---|---|---|
| Definition | Non-coding repetitive DNA sequences mainly found in centromeres and heterochromatin. | Genetic elements that replicate via RNA intermediate and integrate back into the genome. |
| Function | Structurally contributes to chromosome stability and segregation. | Can cause mutations and genome rearrangements; influences genome size. |
| Structure | Highly repetitive tandem repeats, usually short sequences. | Contains coding regions for reverse transcriptase and integrase enzymes. |
| Location | Predominantly centromeric and pericentromeric regions. | Interspersed throughout the genome. |
| Replication Mechanism | Replicates passively with DNA during cell division. | Replicates via "copy-and-paste" mechanism through RNA intermediate. |
| Impact on Genome | Maintains chromosome integrity; minimal sequence variation. | Alters genome structure; contributes to genetic diversity and evolution. |
| Examples | Alpha satellite DNA in humans. | LINEs (Long Interspersed Nuclear Elements), SINEs (Short Interspersed Nuclear Elements). |
Introduction to Satellite DNA and Retrotransposons
Satellite DNA consists of tandemly repeated non-coding sequences that are primarily found in centromeric and pericentromeric regions of eukaryotic chromosomes, playing a crucial role in chromosomal stability and segregation during cell division. Retrotransposons are mobile genetic elements that replicate through an RNA intermediate, integrating copies of themselves into new genomic locations, contributing to genome size variation and evolution. Both satellite DNA and retrotransposons impact genome structure and function but differ fundamentally in their sequence organization and mechanisms of propagation.
Definition and Structural Differences
Satellite DNA consists of tandemly repeated non-coding sequences that are typically found in centromeric and pericentromeric regions of chromosomes, characterized by highly repetitive, short motifs organized in long arrays. Retrotransposons are genetic elements that transpose through an RNA intermediate via a copy-and-paste mechanism, containing coding regions for reverse transcriptase and integrase enzymes, and are dispersed throughout the genome rather than being tandemly repeated. Structurally, satellite DNA lacks protein-coding capacity and forms large, homogeneous blocks, while retrotransposons have distinct functional domains enabling autonomous or non-autonomous mobility within the genome.
Genomic Locations and Distribution
Satellite DNA primarily localizes in centromeric and pericentromeric regions, forming large arrays that contribute to chromosome stability and structure. Retrotransposons are dispersed throughout euchromatic and heterochromatic regions, often interspersed within genes and regulatory elements, impacting genome size and variability. Distribution of satellite DNA is highly tandem and repetitive, whereas retrotransposons exhibit scattered, mobile insertion patterns across the genome.
Mechanisms of Amplification
Satellite DNA amplifies primarily through mechanisms like unequal crossing-over and replication slippage, leading to the expansion of tandemly repeated sequences. Retrotransposons amplify via a copy-and-paste mechanism involving reverse transcription of RNA intermediates, which integrate new copies at different genomic locations. These distinct amplification strategies contribute differently to genome evolution and structural variation in eukaryotic chromosomes.
Evolutionary Origins
Satellite DNA consists of tandemly repeated non-coding sequences often found in centromeric and pericentromeric regions, believed to have evolved through replication slippage and unequal crossing-over mechanisms. Retrotransposons are mobile genetic elements that propagate via an RNA intermediate and reverse transcription, arising from ancient retroviral insertions that contribute to genome plasticity and expansion. The evolutionary origins of satellite DNA and retrotransposons highlight distinct mechanisms of genome evolution, with satellite DNA primarily expanding through local duplication events while retrotransposons spread by transposition across genomic locations.
Functional Roles in the Genome
Satellite DNA primarily contributes to the structural organization of chromosomes, playing a crucial role in centromere function and heterochromatin formation, which ensures proper chromosome segregation during cell division. Retrotransposons influence genome evolution by promoting genetic diversity through their ability to copy and insert themselves into new genomic locations, thereby impacting gene regulation and genome plasticity. Both elements are vital for maintaining genomic stability, but satellite DNA is more associated with chromosomal architecture, while retrotransposons drive dynamic genomic changes.
Impact on Genome Stability
Satellite DNA consists of tandemly repeated non-coding sequences that contribute to chromosomal structure and centromere function, helping maintain genome stability through proper chromosome segregation. Retrotransposons, mobile genetic elements that can replicate and insert themselves into new genomic locations, pose a risk to genome integrity by causing insertional mutations, genomic rearrangements, and chromosomal instability. While satellite DNA supports structural genome stability, retrotransposons drive genomic variability and instability affecting evolutionary processes and disease development.
Detection and Analysis Techniques
Detection of satellite DNA primarily relies on techniques such as fluorescence in situ hybridization (FISH) and Southern blotting, which allow for visualization and size estimation of tandem repeats. Retrotransposons are frequently analyzed using PCR-based methods, next-generation sequencing (NGS), and bioinformatics tools that identify insertion sites and sequence homology. Advanced methods like long-read sequencing enable precise mapping of both satellite DNA arrays and retrotransposon integration, facilitating comprehensive genomic studies.
Relevance in Human Health and Disease
Satellite DNA consists of tandemly repeated, non-coding sequences involved in chromosomal stability and centromere function, with alterations linked to cancer and genetic disorders. Retrotransposons are mobile genetic elements that can disrupt gene function and genome integrity, contributing to diseases like hemophilia, neurodegeneration, and various cancers. Both elements influence genome architecture and epigenetic regulation, making them critical factors in human health and disease susceptibility.
Recent Research and Future Directions
Recent research highlights distinct functional roles of satellite DNA and retrotransposons in genome regulation and stability, with satellite DNA primarily influencing chromosomal architecture and heterochromatin formation, while retrotransposons contribute to genome plasticity and gene expression modulation. Advances in high-throughput sequencing technologies have enabled detailed mapping of satellite DNA arrays and retrotransposon insertion sites, revealing their dynamic interplay in epigenetic regulation and genome evolution. Future research aims to exploit CRISPR-based tools to manipulate these repetitive elements for therapeutic gene editing and to further elucidate their roles in aging, cancer, and neurological disorders.
Satellite DNA Infographic
 libterm.com
libterm.com