Oligogenic traits are influenced by a few genes, distinguishing them from single-gene or polygenic traits. Understanding the complex interactions between these limited genes can provide insights into certain diseases and inherited characteristics. Explore the rest of the article to discover how oligogenic inheritance impacts your genetic makeup.
Table of Comparison
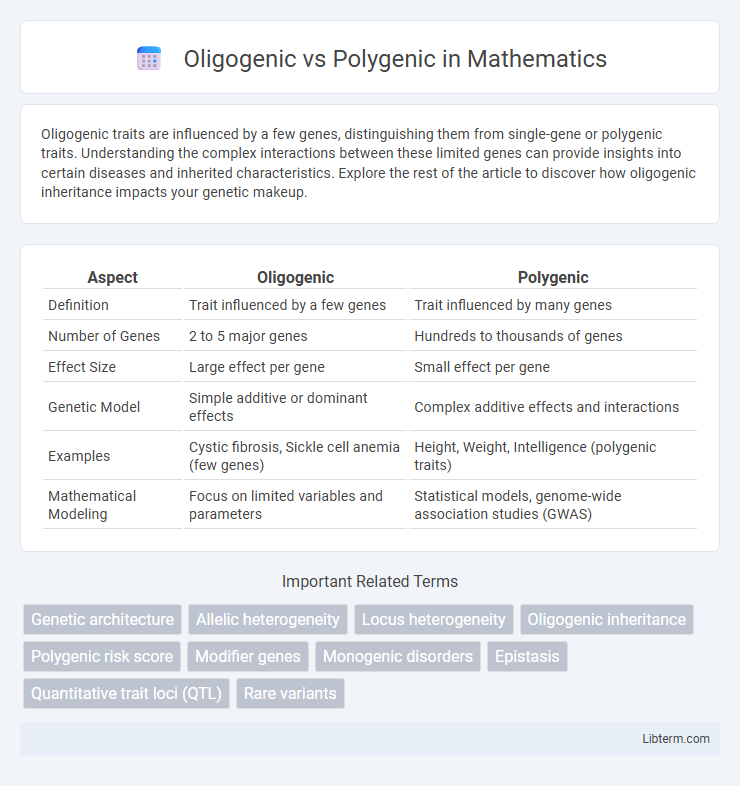
| Aspect | Oligogenic | Polygenic |
|---|---|---|
| Definition | Trait influenced by a few genes | Trait influenced by many genes |
| Number of Genes | 2 to 5 major genes | Hundreds to thousands of genes |
| Effect Size | Large effect per gene | Small effect per gene |
| Genetic Model | Simple additive or dominant effects | Complex additive effects and interactions |
| Examples | Cystic fibrosis, Sickle cell anemia (few genes) | Height, Weight, Intelligence (polygenic traits) |
| Mathematical Modeling | Focus on limited variables and parameters | Statistical models, genome-wide association studies (GWAS) |
Understanding Oligogenic and Polygenic Inheritance
Oligogenic inheritance involves the combined effect of a few genes, typically two to five, each having a moderate impact on a trait or disease, whereas polygenic inheritance results from the cumulative influence of many genes, often dozens or hundreds, each contributing a small effect. Oligogenic traits show a clearer pattern of inheritance with specific gene interactions, while polygenic traits exhibit continuous variation and complex genetic architecture influenced by environmental factors. Understanding the distinction between oligogenic and polygenic inheritance is crucial for genetic research, disease prediction, and personalized medicine approaches.
Key Differences Between Oligogenic and Polygenic Traits
Oligogenic traits are influenced by a few genes, typically two to five, each having a significant effect on the phenotype, while polygenic traits result from the combined action of many genes, often dozens or more, each contributing a small effect. In oligogenic traits, gene interactions often follow Mendelian inheritance patterns, making them easier to trace and predict, whereas polygenic traits exhibit complex inheritance with continuous variation, such as height or skin color. The key difference lies in the number of genes involved and the magnitude of their individual effects, impacting how these traits are studied and understood in genetics and breeding programs.
Genetic Mechanisms Behind Oligogenic Traits
Oligogenic traits arise from the interaction of a few genes, each contributing significantly to the phenotype, contrasting with polygenic traits controlled by many genes with small effects. The genetic mechanisms behind oligogenic traits involve epistasis and gene-gene interactions where specific alleles at different loci combine to produce distinct phenotypic outcomes. These interactions often result in non-Mendelian inheritance patterns, complicating the prediction of trait heritability and expression.
Genetic Architecture of Polygenic Traits
Oligogenic traits are influenced by a few genes with moderate to large effects, whereas polygenic traits arise from the combined impact of many genes, each contributing small effects. The genetic architecture of polygenic traits involves numerous additive genetic variants dispersed throughout the genome, often identified through genome-wide association studies (GWAS). This complex architecture leads to continuous phenotypic variation and is characteristic of most common human traits such as height, intelligence, and susceptibility to complex diseases.
Examples of Oligogenic Disorders
Oligogenic disorders arise from mutations in a few genes, contrasting with polygenic disorders influenced by many genes. Examples of oligogenic disorders include Bardet-Biedl syndrome, caused by mutations in multiple ciliary genes, and Hirschsprung disease, linked to interactions among RET, EDNRB, and other genes. Understanding these gene interactions helps improve diagnosis and personalized treatment strategies.
Common Polygenic Diseases and Conditions
Common polygenic diseases like type 2 diabetes, hypertension, and coronary artery disease result from the combined effects of multiple genes, each contributing a small risk. In contrast, oligogenic diseases involve a few genes with larger individual effects, leading to more defined inheritance patterns. Genome-wide association studies (GWAS) have identified numerous loci associated with polygenic conditions, highlighting the complex genetic architecture behind these prevalent health issues.
Methods for Identifying Oligogenic and Polygenic Variants
Methods for identifying oligogenic variants often involve targeted gene panels, family-based linkage analysis, and rare variant burden testing to detect interactions among a limited set of genes contributing to a trait. In contrast, polygenic risk scores (PRS), genome-wide association studies (GWAS), and large-scale population sequencing are key approaches for quantifying the cumulative effect of numerous common variants across the genome. Advances in machine learning and integrative multi-omics analyses enhance the resolution and accuracy in distinguishing oligogenic from polygenic genetic architectures.
Implications for Disease Prediction and Management
Oligogenic diseases involve mutations in a few genes, allowing more precise genetic screening and targeted therapies, while polygenic diseases result from the combined effects of many genes with small impacts, complicating prediction and personalized treatment. The limited number of gene variants in oligogenic conditions facilitates clearer genotype-phenotype correlations, improving risk assessment accuracy. In contrast, the polygenic architecture requires integrating genome-wide association study (GWAS) data and polygenic risk scores (PRS) for effective disease prediction and management strategies.
Challenges in Oligogenic vs Polygenic Research
Oligogenic research faces the challenge of identifying interactions between a limited number of genes with moderate effects, complicating the determination of their combined influence on phenotypes. Polygenic studies must analyze the cumulative impact of thousands of variants with small effect sizes, requiring large sample sizes and complex statistical models to detect meaningful associations. Both approaches struggle with issues like variant interpretation, gene-gene interactions, and phenotype heterogeneity, but these difficulties manifest differently due to the scale and complexity inherent in oligogenic versus polygenic architectures.
Future Perspectives in Genetic Trait Analysis
Oligogenic and polygenic models are crucial for advancing precision medicine by enabling more accurate identification of gene interactions influencing complex traits. Future perspectives in genetic trait analysis emphasize integrating multi-omics data and machine learning to better decipher the combined effects of a few oligogenic variants versus numerous polygenic contributions. Improved computational frameworks and high-throughput sequencing technologies will enhance predictive power and clinical applications in disease risk assessment and personalized treatments.
Oligogenic Infographic
 libterm.com
libterm.com