Phagotype refers to the classification of bacteria based on their susceptibility to specific bacteriophages, which are viruses that infect bacterial cells. This technique is crucial in epidemiological studies and clinical diagnostics to identify bacterial strains and track infection sources. Discover how phagotyping can enhance your understanding of microbial identification in the rest of this article.
Table of Comparison
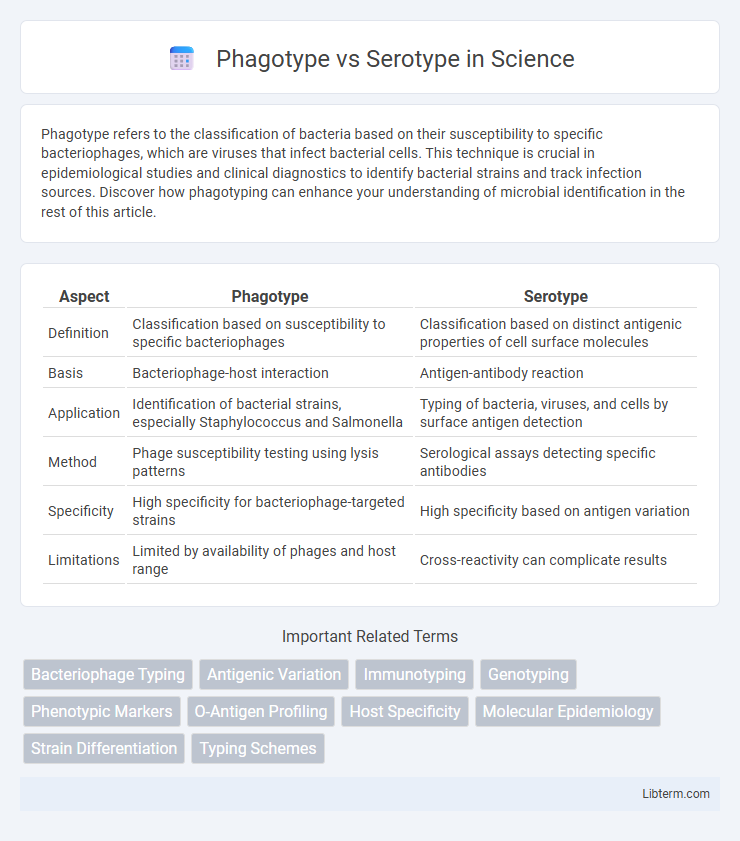
| Aspect | Phagotype | Serotype |
|---|---|---|
| Definition | Classification based on susceptibility to specific bacteriophages | Classification based on distinct antigenic properties of cell surface molecules |
| Basis | Bacteriophage-host interaction | Antigen-antibody reaction |
| Application | Identification of bacterial strains, especially Staphylococcus and Salmonella | Typing of bacteria, viruses, and cells by surface antigen detection |
| Method | Phage susceptibility testing using lysis patterns | Serological assays detecting specific antibodies |
| Specificity | High specificity for bacteriophage-targeted strains | High specificity based on antigen variation |
| Limitations | Limited by availability of phages and host range | Cross-reactivity can complicate results |
Introduction to Phagotype and Serotype
Phagotype refers to the classification of bacteria based on their susceptibility to specific bacteriophages (viruses that infect bacteria), providing precise identification of bacterial strains. Serotype involves the categorization of microorganisms based on distinct surface antigens recognized by antibodies, essential for differentiating species or subspecies within pathogens like Salmonella or Streptococcus. Both phagotyping and serotyping are critical methods in microbiology for epidemiological tracking and infection control.
Definitions: What is a Phagotype?
A phagotype refers to the classification of bacteria based on their susceptibility to infection by specific bacteriophages, which are viruses that infect and lyse bacterial cells. It is a phenotypic method used in microbiology to differentiate bacterial strains by observing patterns of phage-induced lysis. This technique provides critical information for identifying bacterial subtypes in epidemiological studies and bacterial typing.
Understanding Serotype: Key Concepts
Serotype classification is based on the identification of distinct antigenic structures, typically surface proteins or polysaccharides, found on bacterial or viral strains. This method enhances pathogen differentiation by detecting variations in cell surface antigens recognized by specific antibodies, crucial for epidemiological tracking and vaccine development. Serotyping provides detailed insights into strain-specific immune responses, enabling targeted treatment and prevention strategies.
Historical Development of Phagotyping and Serotyping
Phagotyping emerged in the early 20th century as a method to classify bacteria based on their susceptibility to specific bacteriophages, revolutionizing bacterial identification and epidemiology by enabling precise strain differentiation. Serotyping developed concurrently by utilizing antibodies to detect distinct surface antigens on bacteria, which facilitated rapid and reliable classification of pathogenic strains crucial for vaccine development. Both techniques laid foundational roles in microbiology, with phagotyping's precision in bacteriophage susceptibility complementing serotyping's antigenic specificity for advancing infectious disease control.
Methodologies: How Phagotyping is Performed
Phagotyping is performed by exposing bacterial isolates to a set of specific bacteriophages to observe lysis patterns, which helps identify strain-specific susceptibility profiles. In contrast, serotyping involves detecting unique surface antigens on bacteria using specific antibodies through agglutination or immunofluorescence assays. Phagotyping relies on bacteriophage-host interactions, while serotyping depends on antigen-antibody recognition, highlighting distinct methodological approaches for bacterial classification.
Methodologies: How Serotyping is Conducted
Serotyping is conducted by identifying specific surface antigens using agglutination tests with known antisera, allowing precise classification of bacterial strains based on their unique antigenic structures. This method involves mixing bacterial samples with antibody-coated latex beads or specific antisera to observe visible clumping reactions indicative of antigen-antibody binding. Such antigen-based detection contrasts with phagotyping, which relies on susceptibility to bacteriophage infection, making serotyping a direct technique for antigenic characterization in microbial diagnostics.
Applications in Clinical Microbiology
Phagotyping and serotyping are essential tools in clinical microbiology for bacterial strain identification and epidemiological tracking. Phagotyping classifies bacteria based on their susceptibility to specific bacteriophages, aiding in the differentiation of strains within species like Staphylococcus aureus and Salmonella. Serotyping relies on antigen-antibody reactions targeting surface molecules such as O, H, and K antigens, providing vital information for diagnosing infections, vaccine development, and outbreak investigations.
Comparative Advantages and Limitations
Phagotyping enables precise identification of bacterial strains by analyzing their susceptibility to specific bacteriophages, offering high specificity and reproducibility in epidemiological investigations. Serotyping classifies bacteria based on surface antigen variations, providing rapid results and broad applicability across pathogenic species but with lower strain differentiation. Phagotyping is limited by the requirement of suitable phage libraries and complex interpretation, while serotyping may produce cross-reactivity and less resolution in distinguishing closely related strains.
Role in Disease Surveillance and Outbreak Tracking
Phagotype and serotype classification play crucial roles in disease surveillance and outbreak tracking by enabling precise identification of bacterial strains responsible for infections. Phagotyping differentiates bacteria based on susceptibility to specific bacteriophages, while serotyping classifies strains based on distinct surface antigens, particularly in pathogens like Salmonella and Streptococcus. These methods enhance epidemiological investigations by facilitating rapid detection of outbreak sources, tracking transmission pathways, and supporting targeted public health interventions.
Future Trends in Bacterial Typing Techniques
Future trends in bacterial typing techniques emphasize integrating whole-genome sequencing (WGS) with phagotype and serotype data to enhance resolution and accuracy in strain differentiation. Advanced bioinformatics tools leveraging machine learning are increasingly applied to predict bacterial behavior and antibiotic resistance from typing profiles. The shift towards rapid, high-throughput genomic methods is set to replace traditional phenotypic approaches, enabling more precise epidemiological tracking and outbreak management.
Phagotype Infographic
 libterm.com
libterm.com